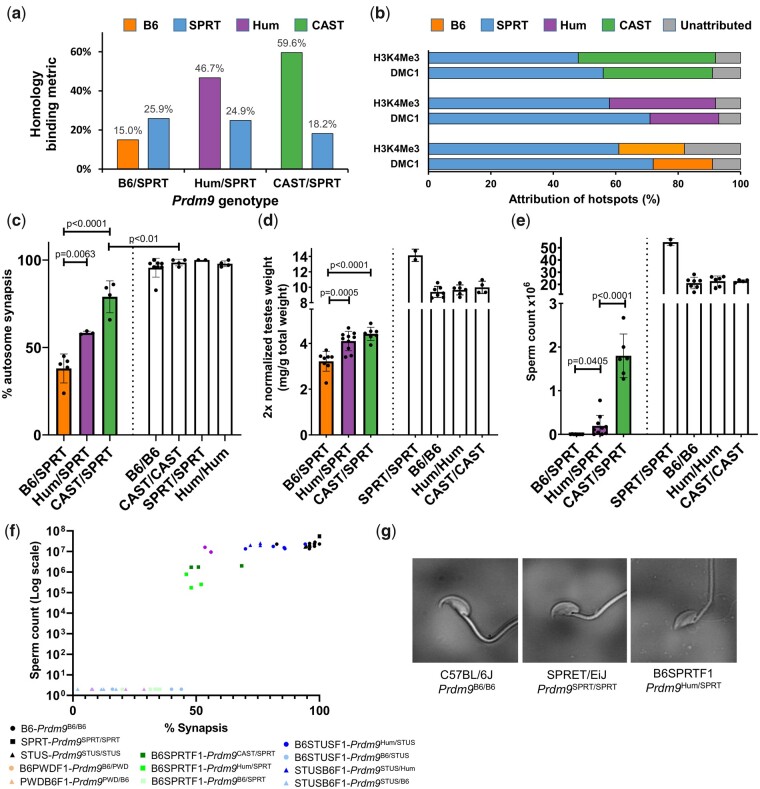
Fig. 2.
The introduction of de novo Prdm9 alleles partially rescues fertility across the species barrier. (a) The proportion of DMC1 SSDS reads originating from matched positions on the two homologs (as defined by B6 contribution between 25% and 75%) for each of the alleles (SPRT: blue, B6: orange, Humanized: purple, CAST: green) present in B6SPRTF1 hybrids harboring the wild-type Prdm9B6 allele (B6/SPRT) or the engineered Prdm9HUM (Hum/SPRT) and Prdm9CAST (CAST/SPRT) alleles. (b) Overall hotspot attribution in these respective hybrids (bottom to top) using DMC1 ssDNA and H3K4Me3 ChIP sequencing peaks, colored as (a) and using gray for unattributed hotspots. (c) Mean proportion of normal autosomal synapsis (n=4), (d) testis weight (B6/SPRT, n=8; Hum/SPRT, n=10; CAST/SPRT, n=7), and (e) total sperm count (B6/SPRT, n=8; Hum/SPRT, n=10; CAST/SPRT, n=6) in the three hybrids and parental controls. Error bars show 1 SD. (f) The synapsis rate as determined by SYCP3/HORMAD2 staining, versus epididymal sperm count, are plotted for individual mice. PWDB6F1 and B6PWDF1 data are from Davies et al. (2016). An arbitrary sperm count of 2 has been given to mice with no sperm to enable the plot. (e) Morphology of representative sperm is shown for wild-type C57BL/6J mice, wild-type SPRET/EiJ mice, and the humanized hybrid, B6SPRTF1.