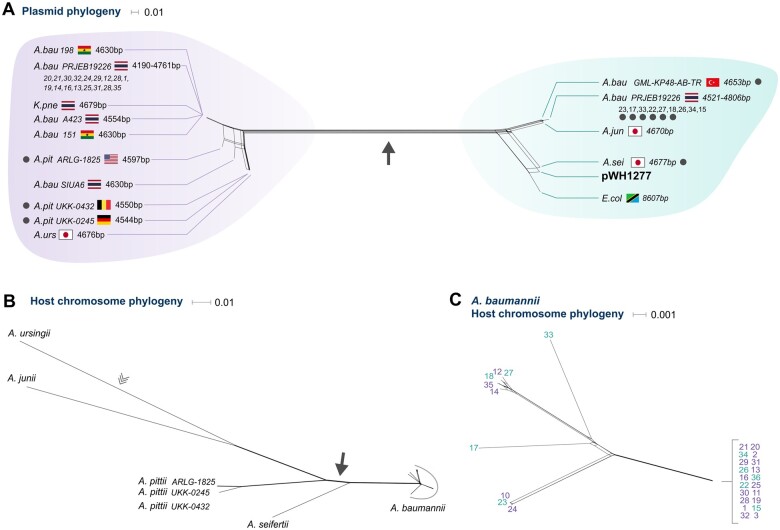
Fig. 4.
Evolutionary reconstruction of plasmid pWH1277. Phylogenetic network of pWH1277 homologous plasmids (A) and chromosome (B, C) of the hosting isolates. (A) The plasmid phylogenetic network was inferred from a multiple alignment of the full-length plasmid genome sequence. For simplicity, closely related Acinetobacter baumannii isolates within the same bioproject are abbreviated with a single label. Strains marked with gray dots were included in the evolutionary rate comparison. Corresponding splits in the plasmid and chromosome phylogenies are marked by a filled arrow. Isolate label includes country of origin and plasmid contig length. We note that the plasmid is not expected to replicate in Escherichia and Klebsiella. Further validation of the host taxonomic classification according to the genomic data (see Materials and Methods) confirmed their reported taxonomy; we consider the observation of pWH1277 in those hosts a rare event. (B) The chromosome phylogenetic network was inferred from a concatenated alignment of 1,617 universal single-copy gene families shared among all Acinetobacter isolates identified as pWH1277-derivative hosts. The root position is marked by an open arrowhead as previously inferred (Mateo-Estrada et al. 2019). (C) Phylogenetic network of A. baumannii isolates was inferred from 2,645 chromosomal universal genes. The color of the labels corresponds to the plasmid groups observed in (A). Plasmids from the purple and blue groups are intermixed in the chromosomal phylogeny, indicating mixed plasmid origins in within A. baumannii. Isolate serial number in (A) and (C) identifies the isolates as in supplementary table 4, Supplementary Material online.