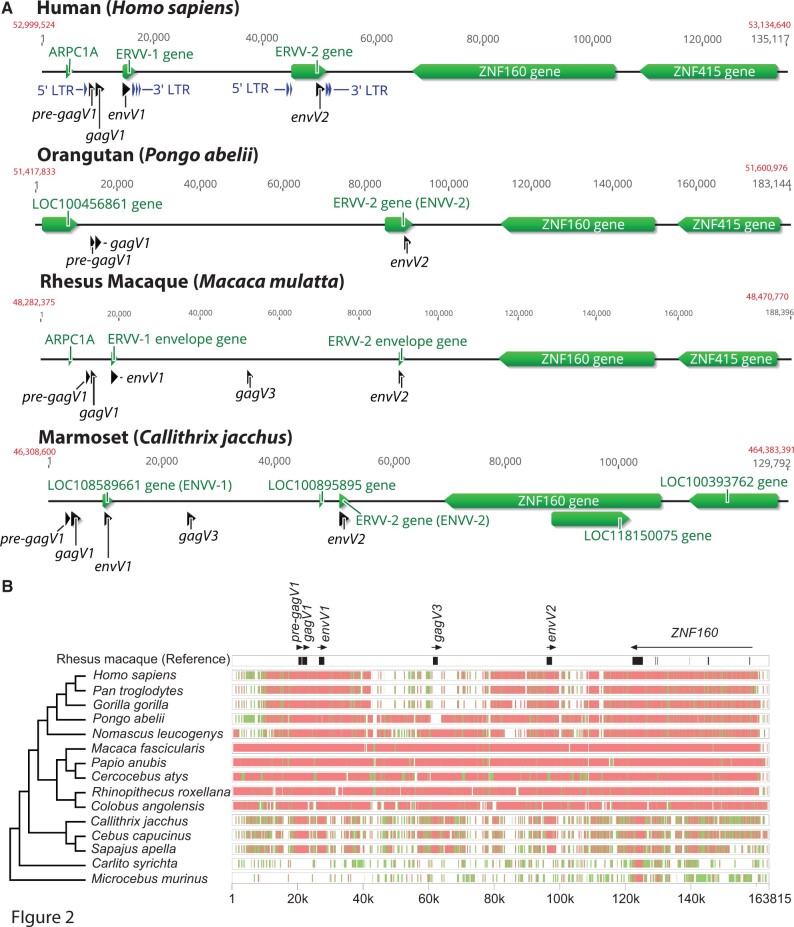
Fig. 2.
Conserved synteny of the HERV-V locus in primates. (A) Chromosomal context of the HERV-V locus is shown for human (GRCh38.p13, chromosome 19), orangutan (Susie_PABv2, chromosome 19), rhesus macaque (Mmul_10, chromosome 19), and marmoset (Callithrix_jacchus_cj1700_1.1, chromosome 22) genomes. Refseq annotated genes are shown in green. Location of the pre-gagV1, gagV1, gagV3, envV1, and envV2 ORFs are indicated in black. Starting and ending coordinates of the depicted region for each species are indicated in red. The figure was created using Geneious (Kearse et al. 2012). (B) Genomic segments containing the ERV-V locus and ZNF160 starting with 20k bases upstream of the pre-gagV1 sequence were extracted from the NCBI genome database for each of the indicated species. Cross-species homologies were analyzed using the MultiPipMaker alignment tool (Elnitski et al. 2010). ORFs of pre-gagV1, gagV1, gagV3, envV1, and envV2 as well as exons of ZNF160 are shown in black boxes in the rhesus macaque reference assembly. Regions with less than 75% and more than 50% identity are shown as green boxes. Regions with more than 75% identity are shown as red boxes. A cladogram representing the phylogenetic relationships between the primate species is shown on the left. The species tree was generated with TimeTree (Hedges et al. 2015).