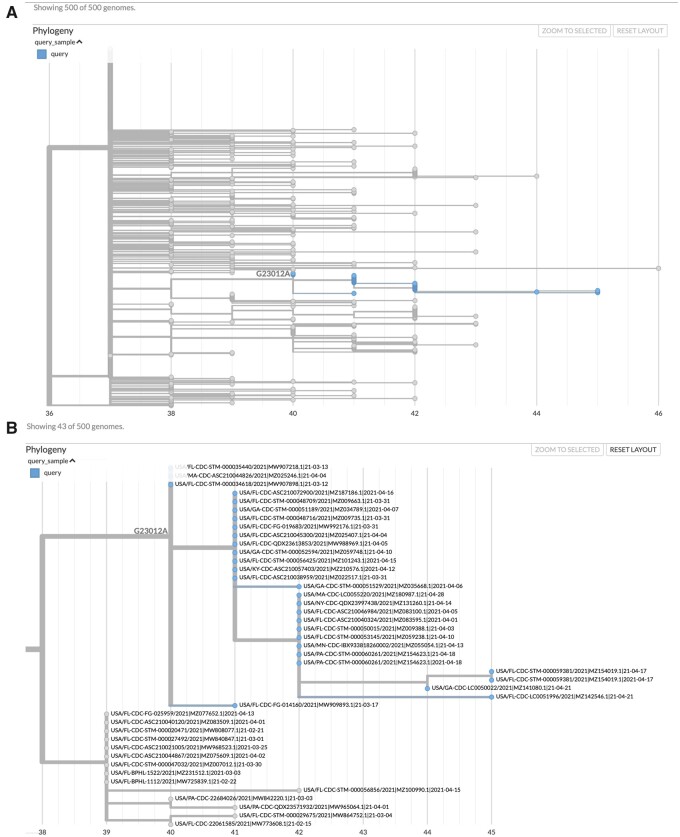
Fig. 2.
matUtils can generate informative visuals with Auspice. The above trees represent a clade of related B.1.1.7 samples from the United States which secondarily acquired the potentially important spike protein mutation E484K, which is caused by the nucleotide mutation G23012A. These trees were obtained by running the command “matUtils extract -i public-2021-06-09.all.masked.nextclade.pangolin.pb.gz -c B.1.1.7 -m G23012A -H ‘(USA.*)’ -N 500 -j clade_trees -d clade_out,” which selects all samples from clade B.1.1.7 which acquired this mutation and are from the United States, then identifies the minimum set of 500 sample subtrees which contain all of these samples, creating an Auspice v2 format JSON for each subtree (Hadfield et al 2018). This results in 35 distinct subtree JSON files of 500 samples each in the output directory. Panel A represents the entirety of subtree six as viewed with Auspice (Hadfield et al 2018), including blue highlights and a branch label where our mutation of interest occurred. Panel B is zoomed in on this subtree and its sister clade; at this scale, we can read individual sample names and observe that this specific strain has been actively spreading in the United States during April 2021.