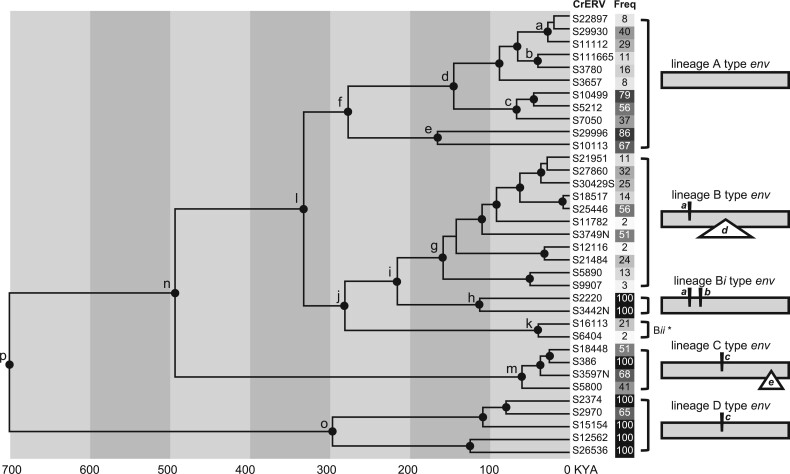
Fig. 2.
Coalescent phylogeny, env structural variation, and population frequency of representative full-length nonrecombinant CrERVs. This is an unrooted phylogeny based on the alignment of 34 CrERV sequences (1,477–8,633 relative to JN592050) with no signature of recombination. The region of the genome spanning a portion of env (6,923–7,503 bp relative to JN592050) was manually blocked to accommodate variable regions in different CrERV lineages. Nodes with at least 95% posterior probability support are marked by black dots. The high posterior density for each labeled node is shown in supplementary table S7, Supplementary Material online. The percentage of mule deer that carry a CrERV is given in a linear gray scale background (white = 0, black = 100) (see supplementary table S6, Supplementary Material online, for additional information). Diagrams on the right side depict the lineage-specific structural variations in the CrERV env. Insertions are represented by italicized lower case characters (a, b, and c) above the sequence; deletions are shown within a triangle under the sequence (d and e). *, a schematic for Lineage Bii could not be made because the five representatives of this lineage had incomplete coverage of env.