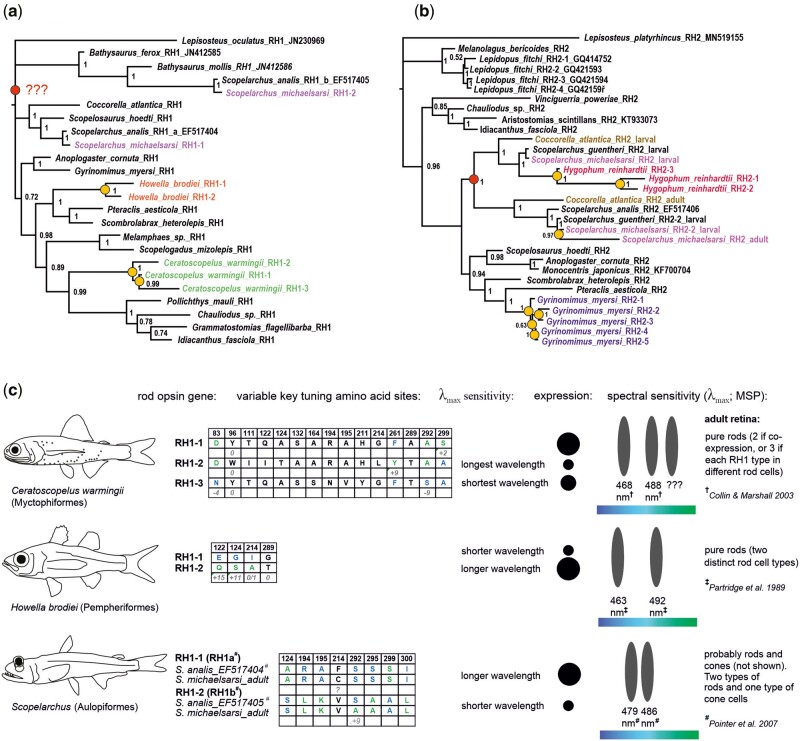
Fig. 3.
Gene trees of the (A) RH1 and (B) RH2 opsin genes found in the retinal transcriptomes of deep-sea fishes. Species with multiple copies are highlighted in color. Additional gene sequences from public databases are listed with their GenBank accession numbers. Note the topology within Aulopiformes; the adult RH2s of Cocorrella atlantica and Scopelarchus cluster together as do the major larval RH2s. Yellow circles mark lineage-specific gene duplication events, while red circles pinpoint the ancestral duplication of RH1 impacting the Scopelarchus genus, and the duplication of RH2 in the aulopiform ancestor (or at least the common ancestor of Coccorella and Scopelarchus). (C) Key tuning spectral site mutations in species with multiple rod opsins and indicative wavelength shifts based on previous in vitro experiments. Known shifts are listed in nanometers if available. Blue and green letters in the tables stand for the shorter- and longer-shifting amino acid variants, respectively. Multiple different rod opsins have been found in three species, Ceratoscopelus warmingii (Myctophiformes), Howella brodiei (Pempheriformes), and Scopelarchus michaelsarsi (Aulopiformes). Note that the RH1 copies in Scopelarchus seem to show a mixed pattern—the longer-wavelength sensitive copy (RH1a; confirmed by in vitro measurements by Pointer et al. (2007) carries also several shorter-shifting amino-acid sites as compared to RH1b). Assignment of the longer and shorter wavelength sensitive photoreceptor to a rhodopsin sequence is marked next to the tables. Rhodopsin gene expression marks dominant (large circles) and less abundant (small circles) copies. For functional interpretation of the rod cells in the visual system we considered microspectrophotometry measurements from # = Pointer et al. (2007), † = Collin and Marshall (2003), and ‡ = Partridge et al. (1989).