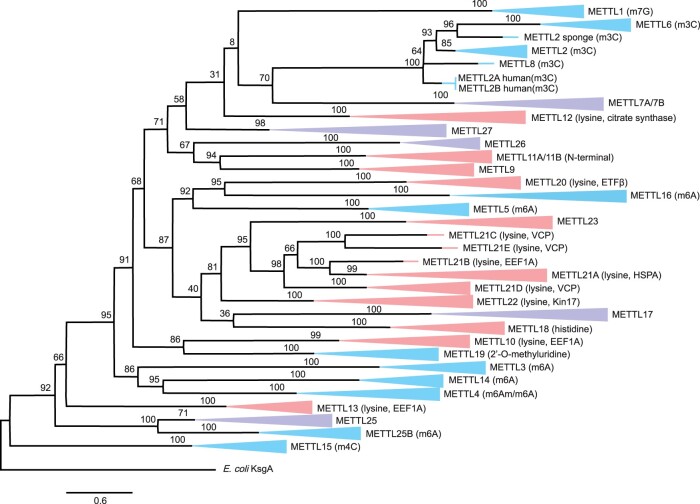
Fig. 2.
Maximum likelihood tree describing the evolutionary relationships across metazoan METTL proteins. Bootstrap values (1,000 replicates) are displayed at each internal node. Branch labels displayed in blue indicate METTLs that have been shown to target DNA or RNA nucleosides, labels in red are METTLs shown to target protein residues, and labels in purple represent METTLs whose function and targets are unknown. The METTL proteins comprise nine different taxa representing eight different phyla: A sponge (Amphimedon queenslandica, Porifera), a stony coral (Acropora millepora, Cnidaria), a sea hare (Aplysia californica, Mollusca), a priapulid (Priapulus caudatus, Priapulida), a shrimp (Litopenaeus vannamei, Arthropoda), a sea star (Acanthaster planci, Echinodermata), an acorn worm (Saccoglossus kowalevskii, Hemichordata), a lancelet (Branchiostoma belcheri, Chordata), and human (Homo sapiens, Chordata). Subtrees with the same METTL gene from multiple taxa have been collapsed for figure readability.