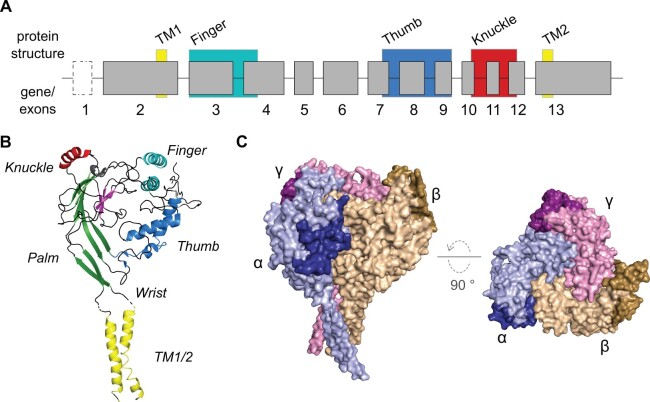
Fig. 1.
Proposed nomenclature of SCNN1 genes and structural features of the human αβγ-ENaC. (A) The SCNN1 genes share a canonical organization in which the coding DNA is distributed over at least 12 exons (exons 2–13). Due to the high variability of exon 2, different predicted start codons located on alternative exons preceding exon 2 as well as the absence of a likely start codon on exon 2 in certain species, make an additional exon(-s) necessary and are therefore depicted in a dashed box. Structural features obtained from the cryo-EM-derived structure of human αβγ-ENaC were imposed to the respective encoding exons. All structural features are highlighted with colored boxes. Only exon sizes and not intron sizes are drawn to scale. (B) SCNN1 proteins share an overall hand-like structure, including regions representing “finger,” “thumb,” “palm,” “wrist,” and “knuckle,” holding a “ball of β-sheets” (shown in magenta). Transmembrane regions are termed TM1 and TM2. The image shows the human ENaC α-subunit (Noreng et al. 2018). (C) Surface model of the cryo-EM-derived structure of human αβγ-ENaC (Noreng et al. 2018). Gating Relief of Inhibition by Proteolysis (GRIP) domains are highlighted in darker colors.