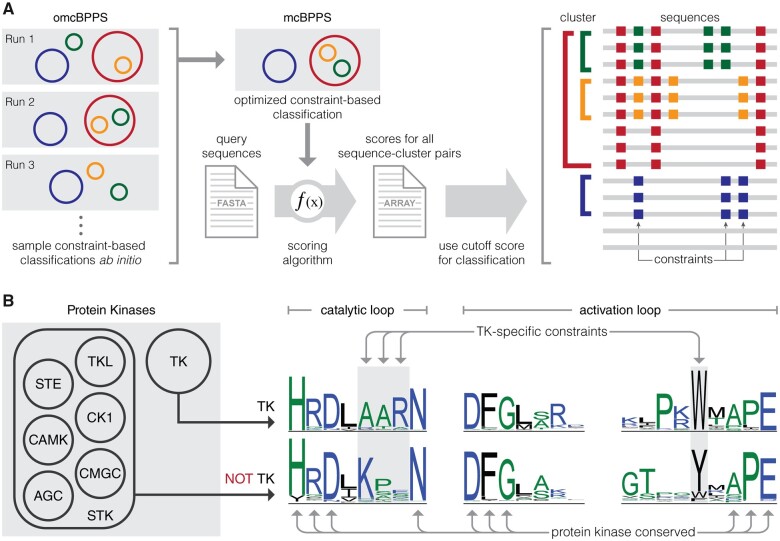
Fig. 1.
Workflow for constraint-based hierarchical clustering of tyrosine kinase sequences. (A) Multiple omcBPPS runs were used to sample various constraint-based hierarchical classifications for tyrosine kinases. An optimal constraint-based classification was determined based on LPR scores calculated using mcBPPS. Next, a scoring algorithm was used to score sequence-cluster pairs and to include or exclude tyrosine kinase sequences from each cluster defined in the optimal classification. An example constraint-based hierarchical classification is shown on the right. Clusters are represented as colored brackets, sequences are represented as gray lines, and constraints specific to each cluster are represented by squares colored according to the cluster to which they belong. For example, sequences in green cluster share sequence constraints denoted by green squares, which are not found in sequences outside of the green cluster, as well as sequence constraints denoted by red squares, which are not found in sequences outside of the red cluster. The last two sequences are not included in any cluster as they lack any of the cluster-specific constraints defined in the constraint-based classification. (B) A visual representation of cluster-specific constraints is shown using previously published data on tyrosine kinase-specific constraints (Mohanty et al. 2016). Seven clusters of protein kinases are shown on the left, where tyrosine kinases are clustered separately from other protein kinases. A sequence logo of tyrosine kinase sequences is shown alongside a sequence logo of all other protein kinases. Sequence motifs such as HRD, DFG, and APE are conserved throughout all protein kinases, whereas the catalytic loop AAR motif and the activation loop tryptophan are defined as tyrosine kinase-specific constraints because their conservation is specific to the tyrosine kinase cluster.