Abstract
Background
Data on household transmission of carbapenemase-producing Enterobacterales (CPE) remain limited. We studied risk of CPE household co-colonization and transmission in Ontario, Canada.
Methods
We enrolled CPE index cases (identified via population-based surveillance from January 2015 to October 2018) and their household contacts. At months 0, 3, 6, 9, and 12, participants provided rectal and groin swabs. Swabs were cultured for CPE until September 2017, when direct polymerase chain reaction (PCR; with culture of specimens if a carbapenemase gene was detected) replaced culture. CPE risk factor data were collected by interview and combined with isolate whole-genome sequencing to determine likelihood of household transmission. Risk factors for household contact colonization were explored using a multivariable logistic regression model with generalized estimating equations.
Results
Ninety-five households with 177 household contacts participated. Sixteen (9%) household contacts in 16 (17%) households were CPE-colonized. Household transmission was confirmed in 3/177 (2%) cases, probable in 2/177 (1%), possible in 9/177 (5%), and unlikely in 2/177 (1%). Household contacts were more likely to be colonized if they were the index case’s spouse (odds ratio [OR], 6.17; 95% confidence interval [CI], 1.05–36.35), if their index case remained CPE-colonized at household enrollment (OR, 7.00; 95% CI, 1.92–25.49), or if they had at least 1 set of specimens processed after direct PCR was introduced (OR, 6.46; 95% CI, 1.52–27.40).
Conclusions
Nine percent of household contacts were CPE-colonized; 3% were a result of household transmission. Hospitals may consider admission screening for patients known to have CPE-colonized household contacts.
Keywords: carbapenem-resistant Enterobacteriaceae, β-lactamases, disease transmission, epidemiology, public health
This prospective cohort study of 95 carbapenemase-producing Enterobacterales (CPE) index cases and 177 household contacts detected 9% of household contacts as CPE-colonized; 3% were due to confirmed/probable household transmission. Hospital admission screening programs may consider including household contacts of known CPE cases.
Carbapenemase-producing Enterobacterales (CPE) are resistant to all β-lactams and many other antibiotics [1, 2]. Invasive infections caused by these organisms are associated with case fatality rates as high as 70% [1, 3–6]. In south-central Ontario, Canada, the incidence of CPE has approximately doubled every 2 years since it was first detected in 2007, to 1.3 clinical isolates per 100 000 population in 2017 [7]. Understanding the epidemiology of CPE transmission is critical to guide prevention and control programs.
Infection control recommendations for CPE prevention have consisted of identifying colonized patients in healthcare facilities and instituting strategies to prevent patient-to-patient spread [8, 9]. Although these measures are necessary, they are insufficient if CPE transmission occurs in the community. There are numerous potential sources of exposure to CPE in community settings [10–12], but data from studies of extended-spectrum β-lactamase–producing Enterobacterales suggest that household transmission may be one important risk [13].
Data regarding the colonization of household contacts of CPE cases are sparse. Publications describe 1 case each in Australia, New Zealand, Belgium, Israel, and the Netherlands [14–18]. We took advantage of population-based surveillance for CPE early in its emergence in south-central Ontario, Canada, to study the risk of and factors associated with colonization of household contacts of CPE cases.
METHODS
Study Design and Setting
The Toronto Invasive Bacterial Diseases Network (TIBDN) has performed population-based surveillance for CPE in metropolitan Toronto and the regional municipality of Peel in Ontario, Canada, since its first detection in the area in October 2007 [19]. Households were eligible for inclusion in this prospective cohort study if a consenting CPE index case identified between January 2015 and October 2018 and at least 1 consenting household contact resided at a fixed address in the community (eg, apartment, house), at which both index case and contact spent at least 20 hours per week. Parents/legal guardians provided consent for participants aged <16 years; participants aged 8–15 years provided assent. Surveillance was approved by the research ethics boards of all TIBDN hospitals. The Sinai Health System Research Ethics Board (Toronto, Canada) approved the study.
Data Collection
At the study’s first home visit (month 0), trained study staff used standardized forms to gather demographic, clinical, personal hygiene, travel, and healthcare exposure data for the prior year from index cases and their household contacts. Follow-up visits occurred at months 3, 6, 9, and 12, at which travel and healthcare exposure in the prior 3 months were recorded. At each home visit, index cases and household contacts provided groin and rectal swabs (Starswabs, Starplex Scientific, Etobicoke, Canada).
Laboratory Procedures
At the microbiology research laboratory at Sinai Health System, swabs were inoculated into 3 mL of brain heart infusion (BHI) broth (Oxoid, Nepean, Canada) and incubated aerobically at 37°C for 24 hours. Broths were subcultured to a biplate of MacConkey crystal violet agar with 2 mg/mL cefpodoxime/0.125 mg/mL meropenem (Oxoid) [20]. One colony of each morphologically distinct oxidase-negative gram-negative bacillus was subcultured to Muller-Hinton agar with a 10-µg meropenem disc (Oxoid). Isolates with meropenem inhibition zones of ≤27 mm were identified to the species level using matrix-assisted laser desorption/ionization time-of-flight (VITEK-MS Plus, bioMérieux, Marcy-l’Étoile, France) [21]. Isolates that were β-CARBA-positive (Bio-Rad, Marnes-la-Coquette, France) had carbapenemase genes identified by polymerase chain reaction (PCR) at the National Microbiology Laboratory (Winnipeg, Canada) or the Public Health Ontario Laboratory (Toronto, Canada) as previously described [22]. From September 2017 onward, carbapenemase genes were detected directly from the incubated BHI broth by PCR given evidence that this was more sensitive than culture [23]. DNA was extracted from 200 µL of incubated BHI broth by boiling, then carbapenemase genes (blaKPC, blaNDM, blaOXA, blaVIM, blaIMP, and blaGES) were detected via multiplex real-time PCR (qPCR) using the ABI 7500 real-time sequence detection system (Applied Biosystems, Foster City, CA) [24]. Culture, as described above, was performed from remaining BHI broth if carbapenemase genes were detected by direct PCR; if an organism was not recovered, another culture method was attempted (Supplementary Methods).
If the household contact and corresponding index case CPE isolate pair were of the same species and carbapenemase gene(s), isolates underwent whole-genome sequencing using NextSeq (Illumina, San Diego, CA) at the National Microbiology Laboratory. Isolates in the index case/household contact pair were conservatively considered highly related if the single nucleotide variant (SNV) distance between them was ≤10 [25, 26], as determined using the SNVPhyl pipeline [27] (Supplementary Methods).
If the household contact and corresponding index case CPE isolate pair had the same carbapenemase gene(s) but were of different species (or the same species with isolates >10 SNVs apart), isolates underwent Illumina and MinION (Oxford Nanopore Technologies, Oxford, UK) long-read sequencing to resolve plasmids. Relatedness of carbapenemase gene–containing plasmids is described for each pair in Supplementary Table 2, including plasmid replicon type, size, and percent identity. For plasmid pairs with ≤99% match, differences and potential relatedness via recombination were noted (Supplementary Methods).
Outcome
The primary outcome was household contact colonization/infection with CPE, defined as at least 1 study or clinical specimen yielding CPE by direct PCR and/or culture.
Definitions
The duration of prestudy exposure of household contacts to each index case was defined as the number of days that the index case spent in the household between their most likely CPE acquisition date and the date of household enrollment and sampling (examples in Supplementary Figure 1).
The time prestudy between index case CPE acquisition, index case identification, and household enrollment meant that not all index cases remained colonized at study start. Index cases were defined as colonized at household enrollment if at least 1 sample yielded CPE at baseline or follow-up visits.
Household transmission was defined as “confirmed” when exposed household contacts had no risk factors for CPE acquisition, an isolate with the same carbapenemase gene as the index case isolate, and either an isolate or carbapenemase gene–containing plasmid with a DNA sequence highly related to that of the index case. Risk factors for CPE acquisition included prior hospitalization, gastrointestinal endoscopy, and travel to the Indian subcontinent (high-risk travel) [19, 28]. Household transmission was “probable” when household contacts had no CPE risk factors and the same carbapenemase gene as the index case, but household contact specimens were PCR-positive only so isolate/plasmid comparisons were unavailable or when the index case and household contact isolates or carbapenemase gene–containing plasmids were highly related and the household contacts had CPE risk factors prior to enrollment but 3 or more negative sets of screening specimens before their first positive. Household transmission was “possible” if household contact and index case isolates had the same carbapenemase genes in isolates or on plasmids that were highly related, but the household contact had high-risk travel concurrent with the index case and 2 or fewer negative sets of screening samples between travel and their first positive. Household transmission was “unlikely” if household contact and index case isolates had different carbapenemase genes or the same carbapenemase genes on unrelated plasmids.
The CPE household acquisition rate per 1000 person-weeks at risk was defined as the number of confirmed, probable, and possible household transmission events divided by the number of weeks household contacts were at risk before and during the study, multiplied by 1000 [13].
Statistical Analyses
Index case and household contact characteristics were summarized using descriptive statistics. Proportions were compared using the χ2 or Fisher exact test as appropriate, and medians were compared using Wilcoxon rank sum tests. Cumulative probabilities of household contacts becoming CPE-colonized were described using Kaplan–Meier survival curves.
To assess risk factors for household contact colonization, we used logistic regression models with generalized estimating equations with an exchangeable correlation matrix to account for clustering at the index case level. First, bivariate analyses were performed to determine whether index case/household contact characteristics were associated with household contact colonization. Significantly associated variables (P < .05) were included in an exploratory multivariable model. Included variables were household contact age (in years), relationship to the index case (spouse vs other), Charlson score [29] (1 or greater vs zero), frequency of primary bathroom sharing with the index case (almost always vs less than almost always), travel to the Indian subcontinent in the year prior to enrollment (yes vs no), number of participating household contacts (2 or more vs 1), whether all home visits occurred prior to the introduction of direct PCR in September 2017 (yes vs no), and whether the index case remained CPE-colonized at household enrollment (yes vs no). Variables were not collinear (variance inflation factors <5). The multivariable model was also run excluding unlikely transmission events. P values <.05 were considered statistically significant using 2-sided tests. Statistical analyses were performed using SAS 9.6M6 (University Edition).
RESULTS
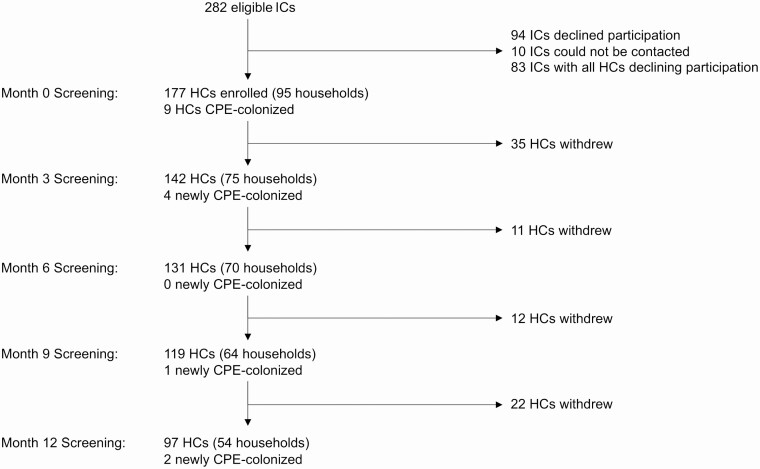
The study flow diagram is shown in Figure 1. Overall, 177 of 343 (52%) household contacts in 95 index case households consented to participate; the median proportion of participating household contacts in these households was 75% (interquartile range [IQR], 25%–100%). The 95 index cases with participating household contacts were similar to the 187 other eligible index cases, except that their CPE acquisition was more likely to be attributed to prior hospitalization abroad (62% vs 39%, P < .0001) and their CPE species was more likely to be Escherichia coli (64% vs 49%, P = .04; Supplementary Table 1).
Figure 1.
Study flow diagram, including number of newly positive HCs at each study visit. Abbreviations: CPE, carbapenemase-producing Enterobacterales; HC, household contact; IC, index case.
Among the 95 participating index cases, the median age was 71 years (range, 3–91) and 37 (39%) were female (Table 1). CPE species among index cases were predominantly E. coli (61, 64%) or Klebsiella pneumoniae (24, 25%), and carbapenemases were primarily NDM- (55, 58%) or OXA-48–like (28, 29%). Index cases had a median of 1 (range, 1–8) participating household contact. The estimated median time of household contact exposure to colonized index cases prior to study start was 97 days (IQR, 51–215). At household enrollment, 61 (64%) index cases remained colonized.
Table 1.
Baseline Characteristics of Participating Index Cases and Household Contacts
| Characteristic | Index Cases (N = 95) |
Household Contacts (N = 177) |
|---|---|---|
| Female sex, n (%) | 37 (39) | 110 (62) |
| Age, median (range), years | 71 (3–91) | 43 (0–95) |
| Charlson score, n (%) | ||
| 0 | 29 (31) | 161 (91) |
| 1–2 | 40 (42) | 15 (8) |
| 3–4 | 15 (16) | 1 (1) |
| ≥5 | 11 (12) | 0 (0) |
| Receipt of antibiotics in prior 6 months, n (%) | 82 (86) | 28 (16) |
| Travel in prior year, n (%) | ||
| Indian subcontinent | 68 (72) | 80 (45) |
| Other countries (except United States/northern Europe) | 11 (12) | 15 (8) |
| None or United States/northern Europe | 16 (17) | 82 (46) |
| Hospitalization in prior year, n (%) | ||
| Abroad | 59 (62) | 3 (2) |
| In Canada | 24 (25) | 13 (7) |
| None | 12 (13) | 161 (91) |
| Index case-specific characteristics | ||
| Number of participating household contacts, median (range) | 1 (1–8) | NA |
| Time from first CPE identification to household enrollment, median (IQR), days | 59 (35–129) | NA |
| CPE species, n (%) | NA | |
| Escherichia coli | 61 (64) | |
| Klebsiella pneumoniae | 24 (25) | |
| Othera | 10 (11) | |
| Carbapenemase produced, n (%) | ||
| NDM | 55 (58) | NA |
| OXA-48–like | 28 (29) | |
| NDM/OXA-48–like | 5 (5) | |
| KPC | 5 (5) | |
| VIM | 2 (2) | |
| Prestudy exposure of household contacts to index cases, median (IQR), days | 97 (51–215) | NA |
| Number of index cases that remained CPE-colonized at enrollment, n (%) | 61 (64) | NA |
| Household contact-specific characteristics | ||
| Relationship to index case, n (%) | ||
| Spouse | NA | 56 (32) |
| Parent | 16 (9) | |
| Child or child-in-law | 60 (34) | |
| Grandchild | 31 (18) | |
| Otherb | 14 (8) |
Abbreviations: CPE, carbapenemase-producing Enterobacterales; IQR, interquartile range; NA, not applicable.
aEnterobacter spp. (7), Providencia rettgeri (1), Morganella morganii (1), Citrobacter freundii (1).
bSibling (4), caregiver (2), child’s parent-in-law (2), cousin (1), fiancé (1), nephew (1), parent-in-law (1), child-in-law’s parent (1), grandchild’s spouse (1).
Among 177 participating household contacts, the median age was 43 years (range, 0–95) and 110 (62%) were female (Table 1). Most household contacts were the spouse (56, 32%) or child/child-in-law (60, 34%) of index cases. At enrollment, 85 (45%) household contacts had at least 1 CPE risk factor other than household exposure: 80 (45%) had traveled to the Indian subcontinent (of whom 3 were hospitalized there and 8 were hospitalized in Canada) and 5 (3%) were only hospitalized in Canada.
Colonization of Household Contacts
Among the 177 participating household contacts, 16 (9%) in 16 (17%) households were CPE-colonized: 9 (5%) were detected at enrollment and 7 (4%) were first detected in follow-up (Figure 1). No household contacts had a clinical specimen that yielded CPE during the study. The median number of positive specimens among colonized household contacts was 2 (interquartile range, 1–2); 8 household contacts had a single positive specimen.
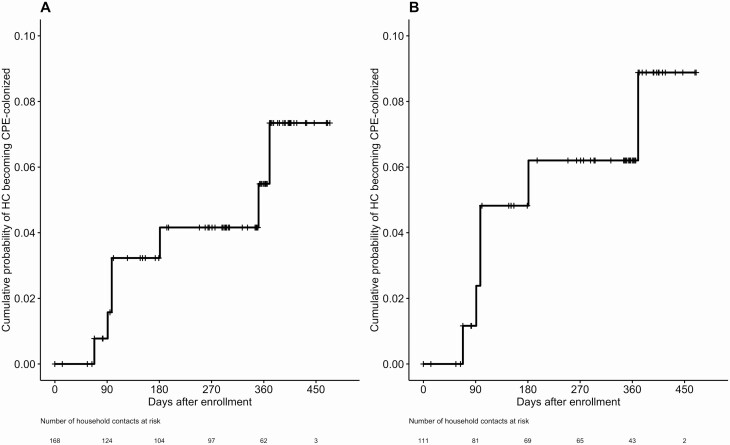
The overall colonization rate was 12/92 (13%) among household contacts with their own risk factors (85 had risk factors at enrollment, 7 had risk factors in follow-up) and 4/85 (5%) among those without risk factors (P = .07). Household contacts who had all home visits prior to the introduction of direct PCR (compared with those who had at least 1 home visit after the introduction of direct PCR) were significantly less likely to be detected as colonized (3/114, 2% vs 13/63, 21%; P < .0001). Among household contacts not colonized at enrollment (n = 168), the cumulative incidence estimate for CPE colonization by 12 months was 5% (Figure 2A). Among the subset of these household contacts whose index cases remained colonized at enrollment (n = 111), CPE colonization by 12 months was 6% (Figure 2B).
Figure 2.
Cumulative incidence of CPE colonization among HCs (A) who were not identified as colonized at enrollment (N = 168) and (B) whose index cases remained colonized at enrollment (N = 111). Month 12 visits (per-protocol visits at 365–395 days after enrollment) were delayed in 14 households due to travel or scheduling challenges, resulting in 33 HCs being censored beyond 395 days. Abbreviations: CPE, carbapenemase-producing Enterobacterales; HC, household contact.
Risk of Household Transmission
Categories of household transmission are shown in Table 2 and Supplementary Table 2. Household transmission was confirmed in 3/177 (2%) household contacts, probable in 2/177 (1%), possible in 9/177 (5%), and unlikely in 2/177 (1%).
Table 2.
Categorization of Likelihood of Transmission From Index Case to Household Contacts in 16 Households With Carbapenemase-Producing Enterobacterales–Colonized Household Contacts
| Transmission Category | HC Risk Factors | IC/HC Species and ST | IC/HC Carbapenemase | Description of Relatedness Between IC and HC Pairs of Isolates/Carbapenemase Gene–Containing Plasmids |
|---|---|---|---|---|
| Confirmed (n = 3) | None | Escherichia coli ST405 | NDM-5 | Highly related isolates (1 SNV) |
| E. coli ST167 | NDM-5 | Highly related blaNDM-5–containing plasmidsa in related isolates (20 SNVs) | ||
| E. coli ST359 | NDM-7 | Highly related blaNDM-7–containing plasmidsa in related isolates (54 SNVs) | ||
| Probable (n = 2) | None | E. coli (IC), culture-negative (HC) | NDM | NA (HC PCR-positive but culture-negative) |
| High-risk co-travel with ICb | Klebsiella pneumoniae ST410 | NDM-1 | Highly related isolates (6 SNVs) | |
| Possible (n = 9) | High-risk co-travel with ICc | E. coli ST372 | OXA-181 | Highly related isolates (0 SNVs) |
| E. coli ST410 | NDM-5 | Highly related isolates (4 SNVs) | ||
| E. coli ST405 | NDM-5 | Highly related isolates (9 SNVs) | ||
| K. pneumoniae ST307 | NDM-5 | Highly related blaNDM-5–containing plasmidsa in related isolates (19 SNVs) | ||
| E. coli ST8655 (IC) and ST940 (HC) | OXA-181 | Highly related blaOXA-181–containing plasmidsa in unrelated isolates (>25 000 SNVs) | ||
| E. coli ST410 (IC) and ST372 (HC) | OXA-181 | Highly related blaOXA-181–containing plasmidsa in unrelated isolates (>25 000 SNVs) | ||
| E. coli ST405 (IC) and ST457 (HC) | NDM-5 | Highly related blaNDM-5–containing plasmidsa in unrelated isolates (>25 000 SNVs) | ||
| E. coli (IC), culture-negative (HC) | OXA | NA (HC PCR positive but culture negative) | ||
| E. coli (IC), isolate unavailable (HC) | NDM | NA (HC isolate unavailable) | ||
| Unlikely (n = 2) | High-risk travel | K. pneumoniae ST14 (IC), E. coli ST8131(HC) | OXA-232 (IC), OXA-181 (HC) | Unrelated plasmidsa |
| K. pneumoniae ST15 (IC), Enterobacter cloacae ST171 (HC) | NDM-1 | Unrelated plasmidsa |
Abbreviations: CPE, carbapenemase-producing Enterobacterales; HC, household contact; IC, index case; NA, not applicable; PCR, polymerase chain reaction; SNV, single nucleotide variant; ST, sequence type.
aSee Supplementary Table 2.
bHC colonization was detected after 4 negative screens post–co-travel with the IC.
cHC colonization was detected after 0 to 2 negative screens post–co-travel with the IC.
All index cases had at least 1 CPE risk factor. In 2 of 3 households with confirmed transmission and 1 of 2 with probable transmission, household contacts were colonized at enrollment (month 0), so transmission occurred prior to study start. In the second household with probable transmission, the household contact and index case had traveled concurrently to the Indian subcontinent; however, the household contact became colonized after 4 consecutive sets of negative screening samples after concurrent travel with the index case.
In the 9 households with possible transmission, household contacts were either colonized at enrollment or had a first positive specimen after only 1–2 sets of negative specimens upon return from concurrent travel with their index cases. Thus, it was not possible to determine whether the household contact acquired CPE from the index case or whether both were exposed during concurrent travel. In the 2 households with unlikely transmission, the household contacts had their own CPE risk factors.
The CPE household acquisition rate was 1.42 per 1000 person-weeks at risk if confirmed, probable, and possible household acquisitions were considered, or 0.50 per 1000 person-weeks at risk if only confirmed and probable household acquisitions were included.
Risk Factors for CPE Colonization of Household Contacts
Table 3 shows index case and household characteristics by whether the household had colonized household contacts, and Table 4 shows household contact characteristics by colonization status. In an exploratory multivariable model, household contacts were significantly more likely to be colonized if they were the index case’s spouse (odds ratio [OR], 6.17; 95% confidence interval [CI], 1.05–36.35), if their index case remained colonized at household enrollment (OR, 7.00; 95% CI, 1.92–25.49), or if at least 1 set of specimens was processed after direct PCR was introduced (OR, 6.46; 95% CI, 1.52–27.40). When unlikely transmission events were excluded, index case spouse (OR, 6.87; 95% CI, 1.07–43.97), index case colonization at household enrollment (OR, 9.80; 95% CI, 1.77–54.33), and direct PCR testing (OR, 5.69; 95% CI, 1.37–23.61) remained associated with household contact colonization.
Table 3.
Index Case and Household Characteristics by Whether Households Had Carbapenemase-Producing Enterobacterales–Colonized Household Contacts
| Characteristic | Households With No CPE-Colonized Household Contacts (n = 79) | Households With at Least 1 CPE-Colonized Household Contacts (n = 16) | P Value |
|---|---|---|---|
| Index case characteristics | |||
| Female sex, n (%) | 33 (42) | 4 (25) | .2 |
| Age, median (IQR), years | 71 (46–77) | 70 (55–78) | .7 |
| Charlson score of 3 or greater, n (%) | 21 (27) | 5 (31) | .7 |
| Receipt of antibiotics in prior 6 months, n (%) | 70 (89) | 12 (75) | .2 |
| Index case remaining CPE-colonized at enrollment, n (%) | 47 (60) | 14 (88) | .04 |
| Travel in prior year (with/without hospitalization), n (%) | |||
| Indian subcontinent | 53 (67) | 15 (94) | .07 |
| Other countries (except United States/Northern Europe) | 10 (13) | 1 (6) | |
| None or United States/Northern Europe | 16 (20) | 0 (0) | |
| Hospitalization in prior year, n (%) | |||
| Outside Canada | 50 (63) | 9 (56) | .6 |
| In Canada | 21 (27) | 4 (24) | |
| None | 8 (10) | 3 (19) | |
| CPE species, n (%) | |||
| Escherichia coli | 51 (65) | 10 (63) | .7 |
| Klebsiella pneumoniae | 19 (24) | 5 (31) | |
| Other | 9 (11) | 1 (6) | |
| Carbapenemase produced, n (%) | |||
| NDM | 47 (59) | 8 (50) | .6 |
| OXA-48–like | 21 (27) | 7 (44) | |
| NDM/OXA-48–like | 4 (5) | 1 (6) | |
| KPC | 5 (6) | 0 (0) | |
| VIM | 2 (3) | 0 (0) | |
| Household characteristics | |||
| Prestudy exposure to index case, median (IQR), days | 97 (51–225) | 86 (52–202) | .9 |
| 2 or more participating household contacts, n (%) | 28 (35) | 5 (31) | .7 |
Abbreviations: CPE, carbapenemase-producing Enterobacterales; IQR, interquartile range.
Table 4.
Household Contact Characteristics by Household Contact Colonization Status
| Characteristic | CPE-Negative Household Contacts (n = 161) | CPE-Colonized Household Contacts (n = 16) | P Value |
|---|---|---|---|
| Female sex, n (%) | 98 (61) | 12 (75) | .4 |
| Age, median (IQR), years | 41 (23–61) | 68 (40–78) | .001 |
| Charlson score of 1 or greater, n (%) | 12 (7) | 4 (25) | .04 |
| Relationship to index case, n (%) | |||
| Spouse | 42 (26) | 14 (88) | <.0001 |
| Child | 44 (27) | 2 (13) | |
| Other | 75 (47) | 0 (0) | |
| Receipt of antibiotics in prior 6 months, n (%)a | 27 (19) | 3 (19) | 1.0 |
| Travel in prior year (with/without hospitalization), n (%) | |||
| Indian subcontinent | 68 (42) | 12 (75) | .04 |
| Other countries except United States/northern Europe) | 14 (9) | 1 (6) | |
| None or United States/northern Europe | 79 (49) | 3 (19) | |
| Hospitalization in prior year, n (%) | |||
| Outside Canada | 3 (2) | 0 (0) | .8 |
| In Canada | 12 (7) | 1 (6) | |
| None | 146 (91) | 15 (94) | |
| Contact with index case, n (%) | |||
| Skin-to-skin contact almost daily | 61 (38) | 6 (38) | 1.0 |
| Always share primary bathroom | 75 (47) | 14 (88) | .002 |
| Always share hand/body towels | 39 (24) | 7 (44) | .09 |
| All home visits prior to introduction of direct polymerase chain reaction, n (%) | 111 (69) | 3 (19) | .0001 |
Abbreviations: CPE, carbapenemase-producing Enterobacterales; IQR, interquartile range.
aData missing for 22 household contacts, as this question was not added to the data collection form until December 2015.
DISCUSSION
In this cohort in Ontario, Canada, 9% of household contacts of CPE cases were identified as CPE-colonized; some were probably from household transmission and others possibly following coexposure with the index case abroad. Household contacts were more likely to be colonized if they were the index case’s spouse, if their index case remained colonized at household enrollment, or if at least 1 home visit occurred after direct PCR was introduced.
No published studies have systematically examined CPE household transmission. However, a recent meta-analysis examined risk of cocarriage/acquisition of extended spectrum β-lactamase–producing Enterobacterales (ESBLE) in households [13]. Among the 7 prospective cohort studies that we identified (6 included in the meta-analysis) [30–36], the proportion of colonized household contacts ranged from 8% to 37%. In 3 of these studies, the rate of ESBLE household acquisition among household contacts could be calculated [13]. Rates in 2 studies were similar to our CPE household acquisition rate of 1.42 per 1000 person-weeks (2.90 and 1.69 acquisitions per 1000 person-weeks at risk) [30, 35]; 1 study of antibiotic exposure in households reported a much higher rate of 19.2 per 1000 person-weeks [36]. CPE and ESBLE may have similar household transmission rates; our estimated rate may be lower than others given our use of whole-genome sequencing to assess transmission.
There are challenges in assessing the risk of CPE household transmission. First, transmission risk presumably starts when a colonized index case and contact share a household and continues until the index case clears colonization. However, the precise date of CPE acquisition among index cases is often unknown, and with sampling every 3 months in our study and others, the precise date of CPE clearance may also be unavailable. Second, a substantial fraction of household contacts may themselves have risk factors for CPE acquisition; as seen in this study, co-acquisition from another source is difficult to distinguish from household transmission. Third, a single negative set of specimens is insufficient to establish absence of colonization [37]. In 1 study [18], which reported household transmission when a household contact was detected as colonized following 1 negative specimen after co-traveling with their index case, their first specimen post-travel may have been a false-negative, and the household contact may have acquired CPE abroad. Finally, direct PCR from patient specimens appears more sensitive than culture for CPE detection [23, 38, 39]. In our study, household contacts were more likely to be detected as colonized if at least 1 study visit occurred after the introduction of direct PCR. We may have missed household transmission events when using culture alone, prior to direct PCR implementation.
This study had limitations. CPE yield may have been improved, and more household transmission detected, if additional body sites had been sampled (eg, urine) and if direct PCR had been used from study start. Nonetheless, our use of overnight broth enrichment and eventual use of direct PCR likely improved yield over other methods. Two household contact specimens had carbapenemase genes identified by direct PCR but did not yield CPE on culture; this limited interpretation of transmission because sequencing could not be performed. It is possible that some of the 8 colonized household contacts with only 1 positive specimen had false-positive PCR results; however, all genes matched their index case genes, making this unlikely. Our index cases primarily had NDM- and/or OXA-48–like producing E. coli; this was expected given frequent travel of our population to the Indian subcontinent, but our results may not be generalizable to populations where other CPE are dominant. Epidemiological data were collected by interviews, and participants’ memories may not be accurate. Our assessment of risk factors for household contact colonization is exploratory because of the small sample size, and we did not consider the role of the household environment in transmission. Risk factors for household acquisition specifically require investigation.
In summary, in this prospective cohort study of CPE index cases and their households, we found that 9% of household contacts were CPE-colonized due to a combination of household transmission and shared risk factors with index cases. This suggests limited spread of CPE in the community and supports other data demonstrating that CPE transmission in high-income countries occurs predominately in hospitals [40]. In light of our findings, hospital CPE admission screening programs may consider including household contacts of known CPE cases (screening is cost-effective in low-prevalence populations [41]), and physicians who treat gram-negative sepsis may select antibiotics differently for household contacts of known CPE cases. There is also an urgent need to improve implementation of CPE prevention and control programs in Ontario [8] and likely other jurisdictions. This has the potential to improve the rate of detection of CPE in our hospitals and subsequently reduce spread [42].
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the infection prevention and control practitioners and clinical microbiology laboratory technologists at Toronto Invasive Bacterial Diseases Network hospitals, the National Microbiology Laboratory (NML) Genomics Core Facility, and Romeo Hizon (NML) and Ken Fakharuddin (NML) for expert technical assistance.
Financial support. This work was supported by the Canadian Institutes of Health Research (grant number 313039). A. J. J. is supported by the Government of Canada Vanier Canada Graduate Scholarship.
Potential conflicts of interest. A. J. M. reports grants from the Canadian Institutes for Health Research during the conduct of the study and grants from Oakville Stamping and Bending and from Merck outside the submitted work. S. P. reports honoraria related to advisory board meetings from Verity, Cipher, Paladin Labs, and Merck; travel reimbursement fees from Merck and Copan; and research support from Accelerate Diagnostics and bioMérieux outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase producing Enterobacteriaceae. Emerg Infect Dis 2011; 17:1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kontopidou F, Giamarellou H, Katerelos P, et al. ; Group for the Study of KPC-Producing Klebsiella pneumoniae Infections in Intensive Care Units. Infections caused by carbapenem-resistant Klebsiella pneumoniae among patients in intensive care units in Greece: a multi-centre study on clinical outcome and therapeutic options. Clin Microbiol Infect 2014; 20:O117–23. [DOI] [PubMed] [Google Scholar]
- 3. Borer A, Saidel-Odes L, Riesenberg K, et al. . Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol 2009; 30:972–6. [DOI] [PubMed] [Google Scholar]
- 4. Souli M, Galani I, Antoniadou A, et al. . An outbreak of infection due to beta-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis 2010; 50:364–73. [DOI] [PubMed] [Google Scholar]
- 5. Kontopoulou K, Protonotariou E, Vasilakos K, et al. . Hospital outbreak caused by Klebsiella pneumoniae producing KPC-2 beta-lactamase resistant to colistin. J Hosp Infect 2010; 76:70–3. [DOI] [PubMed] [Google Scholar]
- 6. Tumbarello M, Viale P, Viscoli C, et al. . Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis 2012; 55:943–50. [DOI] [PubMed] [Google Scholar]
- 7. Zhong Z, Faheem A, Farooqi L, et al. . Emergence of carbapenemase producing Enterobacteriaceae in south central Ontario, Canada. Open Forum Infect Dis 2018; 5(Suppl 1):S365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jamal AJ, Garcia-Jeldes F, Baqi M, et al. ; CPE Investigators of the Toronto Invasive Bacterial Diseases Network . Infection prevention and control practices related to carbapenemase-producing Enterobacteriaceae (CPE) in acute-care hospitals in Ontario, Canada. Infect Control Hosp Epidemiol 2019; 40:1006–12. [DOI] [PubMed] [Google Scholar]
- 9. Provincial Infectious Diseases Advisory Committee. Screening, testing and surveillance for antibiotic-resistant organisms (AROs) in all health care settings. 2013. Available at: https://www.publichealthontario.ca/en/eRepository/PIDAC-IPC_Annex_A_Screening_Testing_Surveillance_AROs_2013.pdf. Accessed 1 May 2020.
- 10. Roschanski N, Guenther S, Vu TTT, et al. . VIM-1 carbapenemase-producing Escherichia coli isolated from retail seafood, Germany 2016. Euro Surveill 2017; 22:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 2011; 11:355–62. [DOI] [PubMed] [Google Scholar]
- 12. Köck R, Daniels-Haardt I, Becker K, et al. . Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect 2018; 24:1241–50. [DOI] [PubMed] [Google Scholar]
- 13. Martischang R, Riccio ME, Abbas M, Stewardson AJ, Kluytmans JAJW, Harbarth S. Household carriage and acquisition of extended-spectrum β-lactamase-producing Enterobacteriaceae: a systematic review. Infect Control Hosp Epidemiol 2020; 41:286–94. [DOI] [PubMed] [Google Scholar]
- 14. Gottesman T, Agmon O, Shwartz O, Dan M. Household transmission of carbapenemase-producing Klebsiella pneumoniae. Emerg Infect Dis 2008; 14:859–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blakiston MR, He H, Roberts SA, Joshua T. Household transmission of NDM-producing E. coli in New Zealand. N Z Med J 2017; 130:63–5. [PubMed] [Google Scholar]
- 16. Blyth CC, Pereira L, Goire N. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae in an Australian child who had not travelled overseas. Med J Aust 2014; 200:386. [DOI] [PubMed] [Google Scholar]
- 17. Yusuf E, Huang TD, Schallier A, et al. . OXA-48 producing Klebsiella pneumoniae in a household contact of a previously infected patient: person-to-person transmission or coincidental community acquisition? Microb Drug Resist 2016; 22:134–6. [DOI] [PubMed] [Google Scholar]
- 18. van Hattem JM, Arcilla MS, Bootsma MC, et al. . Prolonged carriage and potential onward transmission of carbapenemase-producing Enterobacteriaceae in Dutch travelers. Future Microbiol 2016; 11:857–64. [DOI] [PubMed] [Google Scholar]
- 19. Kohler PP, Melano RG, Patel SN, et al. . Emergence of carbapenemase-producing Enterobacteriaceae, south-central Ontario, Canada. Emerg Infect Dis 2018; 24:1674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. European Society of Clinical Microbiology and Infectious Diseases–European Committee on Antimicrobial Susceptibility Testing. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. 2017. Available at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf. Accessed 1 May 2020.
- 21. European Society of Clinical Microbiology and Infectious Diseases - European Committee on Antimicrobial Susceptibility. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. 2013. Available at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf. Accessed 1 May 2020.
- 22. Mataseje LF, Abdesselam K, Vachon J, et al. . Results from the Canadian Nosocomial Infection Surveillance Program on Carbapenemase-producing Enterobacteriaceae, 2010 to 2014 on behalf of the Canadian Nosocomial Infection Surveillance Program. Antimicrob Agents Chemother 2016; 60:1359–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li A, Paterson A, Armstrong A, et al. . PCR versus culture for the detection of carbapenemase-producing Enterobacteriaceae (CPE) in patient and environmental samples [abstract P2316]. In: Program and abstracts of the 28th European Congress of Clinical Microbiology and Infectious Diseases. Madrid, Spain: European Society of Clinical Microbiology and Infectious Diseases, 2018. [Google Scholar]
- 24. Fattouh R, Tijet N, McGeer A, Poutanen SM, Melano RG, Patel SN. What is the appropriate meropenem MIC for screening of carbapenemase-producing Enterobacteriaceae in low-prevalence settings? Antimicrob Agents Chemother 2015; 60:1556–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roer L, Hansen F, Thomsen MCF, et al. . WGS-based surveillance of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. J Antimicrob Chemother 2017; 72:1922–9. [DOI] [PubMed] [Google Scholar]
- 26. Smit PW, Stoesser N, Pol S, et al. . Transmission dynamics of hyper-endemic multi-drug resistant Klebsiella pneumoniae in a Southeast Asian neonatal unit: a longitudinal study with whole genome sequencing. Front Microbiol 2018; 9:1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petkau A, Mabon P, Sieffert C, et al. . SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb Genom 2017; 3:e000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van der Bij AK, Pitout JD. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother 2012; 67:2090–100. [DOI] [PubMed] [Google Scholar]
- 29. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 30. Tandé D, Boisramé-Gastrin S, Münck MR, et al. . Intrafamilial transmission of extended-spectrum-beta-lactamase-producing Escherichia coli and Salmonella enterica Babelsberg among the families of internationally adopted children. J Antimicrob Chemother 2010; 65:859–65. [DOI] [PubMed] [Google Scholar]
- 31. Hilty M, Betsch BY, Bögli-Stuber K, et al. . Transmission dynamics of extended-spectrum β-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis 2012; 55:967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Löhr IH, Rettedal S, Natås OB, Naseer U, Oymar K, Sundsfjord A. Long-term faecal carriage in infants and intra-household transmission of CTX-M-15-producing Klebsiella pneumoniae following a nosocomial outbreak. J Antimicrob Chemother 2013; 68:1043–8. [DOI] [PubMed] [Google Scholar]
- 33. Strenger V, Feierl G, Resch B, et al. . Fecal carriage and intrafamilial spread of extended-spectrum β-lactamase-producing Enterobacteriaceae following colonization at the neonatal ICU. Pediatr Crit Care Med 2013; 14:157–63. [DOI] [PubMed] [Google Scholar]
- 34. Arcilla MS, van Hattem JM, Haverkate MR, et al. . Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis 2017; 17:78–85. [DOI] [PubMed] [Google Scholar]
- 35. Haverkate MR, Platteel TN, Fluit AC, et al. . Quantifying within-household transmission of extended-spectrum β-lactamase-producing bacteria. Clin Microbiol Infect 2017; 23:46.e1–e7. [DOI] [PubMed] [Google Scholar]
- 36. Stewardson AJ, Vervoort J, Adriaenssens N, et al. ; SATURN WP1 Study Group; SATURN WP3 Study Group . Effect of outpatient antibiotics for urinary tract infections on antimicrobial resistance among commensal Enterobacteriaceae: a multinational prospective cohort study. Clin Microbiol Infect 2018; 24:972–9. [DOI] [PubMed] [Google Scholar]
- 37. Feldman N, Adler A, Molshatzki N, et al. . Gastrointestinal colonization by KPC-producing Klebsiella pneumoniae following hospital discharge: duration of carriage and risk factors for persistent carriage. Clin Microbiol Infect 2013; 19:E190–6. [DOI] [PubMed] [Google Scholar]
- 38. Schechner V, Straus-Robinson K, Schwartz D, et al. . Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J Clin Microbiol 2009; 47:3261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singh K, Mangold KA, Wyant K, et al. . Rectal screening for Klebsiella pneumoniae carbapenemases: comparison of real-time PCR and culture using two selective screening agar plates. J Clin Microbiol 2012; 50:2596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. David S, Reuter S, Harris SR, et al. ; EuSCAPE Working Group; ESGEM Study Group . Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 2019; 4:1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lapointe-Shaw L, Voruganti T, Kohler P, Thein HH, Sander B, McGeer A. Cost-effectiveness analysis of universal screening for carbapenemase-producing Enterobacteriaceae in hospital inpatients. Eur J Clin Microbiol Infect Dis 2017; 36:1047–55. [DOI] [PubMed] [Google Scholar]
- 42. Bartsch SM, Wong KF, Stokes-Cawley OJ, et al. . Knowing more of the iceberg: how detecting a greater proportion of carbapenem-resistant Enterobacteriaceae carriers influence transmission. J Infect Dis 2019; 221:1782–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.