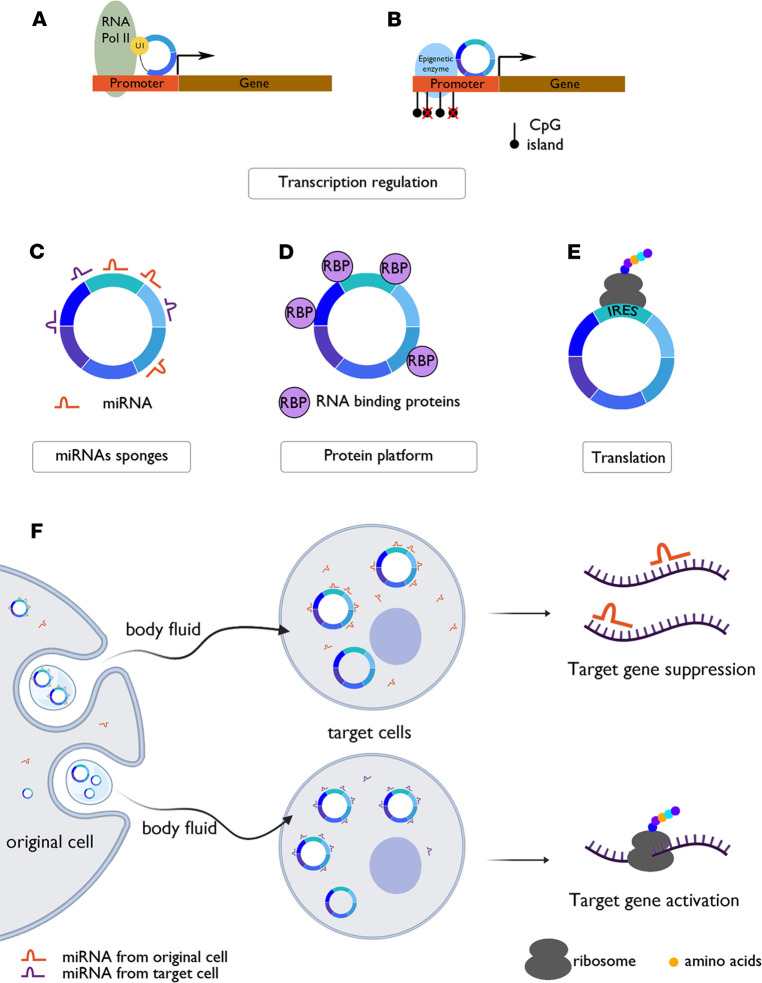
Figure 2. CircRNA functions.
(A) Splicing competitors between back-splicing for circRNAs and canonical splicing for linear RNAs. Multiple circRNAs and linear RNAs can be generated from a single gene locus. Indeed, RNA pairing formed across flanking introns promotes back-splicing, leading to the formation of a circRNA and a linear RNA with exon exclusion, whereas RNA pairing formed within one individual intron promotes canonical splicing, inducing a linear RNA with exon exclusion but not back-splicing. Thus, the competition between these reverse complementary sequences can lead to multiple circRNAs. (B) Transcription regulators. CircRNAs can interact with RNA pol II, the complex containing U1 snRNP, and enhance gene transcription. CircRNAs can also silence genes by regulating parental genes through epigenetic mechanisms. (C) MiRNA sponges. CircRNAs can bind to miRNAs, thus inhibiting miRNA-mRNA conjugation. (D) Protein sponges. CircRNAs can restrain RBP from binding to miRNA or mRNA, thus indirectly regulating the protein biogenesis and the function of RBP. (E) Protein templates. CircRNAs can be translated into peptides when equipped with IRESs and AUG site(s). (F) Packaged into exosomes. Exo-circRNAs are circRNAs in exosomes where they can bind to miRNAs (top). After entering target cells, miRNAs are released and target genes can be silenced. In contrast, when exo-circRNAs are not bound to miRNAs in exosomes (bottom), they are able to sponge specific miRNAs in target cells and target genes are activated. U1, U1 small nuclear ribonucleoprotein particle (snRNP); RNA pol II, RNA polymerase II; RBP, RNA-binding protein; IRES, internal ribosome entry sites.