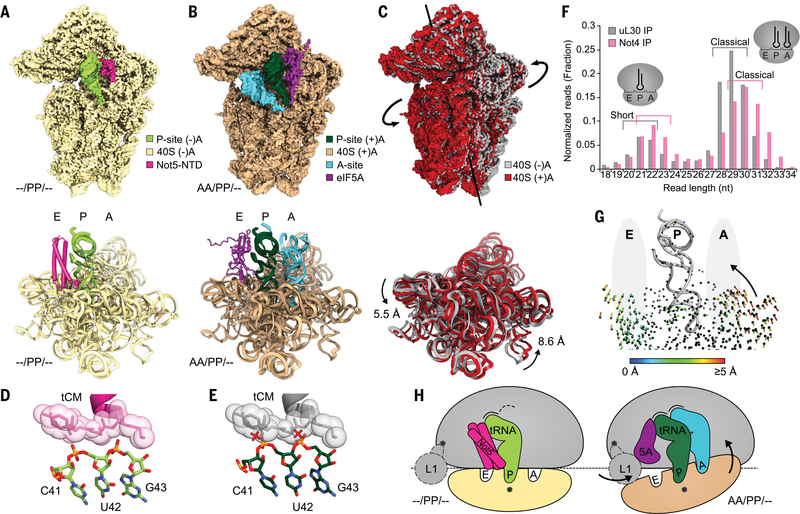
Fig. 4. eIF5A and Not5 compete for the E-site in an A-site–dependent manner.
(A) Density (top) and model (bottom) of the Not5-bound 40S subunit of the (–/PP/–) 80S ribosome (only 18S rRNA model shown). (B) Density (top) and model (bottom) of the eIF5A-bound 40S subunit of the (AA/PP/–) 80S ribosome (only 18S rRNA model shown). (C) Superposition of the densities (top) and the models (bottom) from (A) and (B). Subunit rolling is indicated. Structures in (A) to (C) were aligned with respect to the 60S subunits. (D) The tCM of Not5 is contacted by the anticodon stem of the P-site tRNA in the (–/PP/–) situation (also see Fig. 3D). (E) Conformational changes of the P-site tRNA upon A-site accommodation would cause clashes (red crosses). (F) Length distribution of mRNA fragments during selective ribosome profiling using Not4 and uL30 as baits. (G) Illustration of A-site–E-site coupling. Arrows represent quantified trajectories of Ca atoms (protein) and P atoms (RNA) during transition from (–/PP/–) to (AA/PP/–) conformations. Ribosomal A- and E-sites are schematically indicated with gray ellipsoids. (H) Schematic summary of subunit rolling and the competition between eIF5A and Not5.