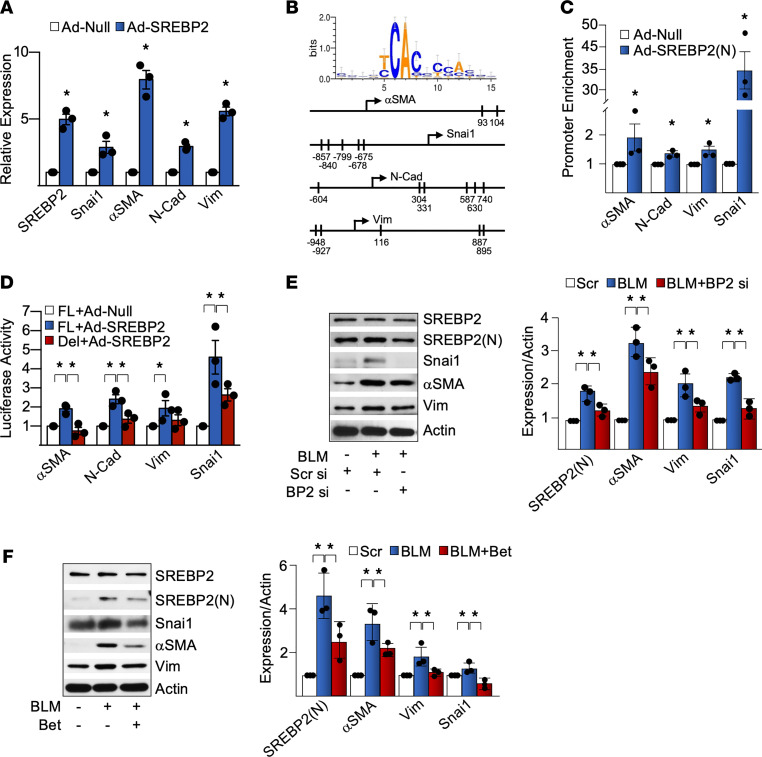
Figure 2. SREBP2 transactivates mesenchymal genes.
(A) HUVECs were infected with Ad-SREBP2(N) or empty vector (Ad-null). The levels of mRNA of the indicated genes were measured by qPCR. (B) Depiction of the predicted sterol regulatory elements (SREs) in the promoter regions of snail family transcriptional repressor 1 (snai1), α-smooth muscle actin (αSMA), N-Cadherin (N-Cad), and vimentin (Vim). (C and D) HUVECs were infected with Ad-SREBP2(N) or empty vector (Ad-Null). (C) Chromatin immunoprecipitation (ChIP) assays were performed on the promoter region of SNAI1 (encoding snai1), ACTA2 (encoding αSMA), CDH2 (encoding N-Cad), and VIM (encoding Vim). (D) Luciferase activity was measured and normalized to that of Renilla. FL, full length promoter; Del, SRE site deletion. (E) HUVECs were transfected with SREBP2 or scrambled control siRNA (20 nM) for 16 hours prior to stimulation with bleomycin (BLM) for an additional 48 hours. (F) HUVECs were pretreated with betulin (Bet) (6 μg/mL) for 2 hours, followed by BLM treatment for an additional 72 hours. The expression of the indicated proteins was measured by Western blot. Data in A and C were analyzed by 2-tailed Student’s t test; data are represented as mean ± SEM from 3 independent experiments (n = 3). Data in D–F were analyzed by 1-way ANOVA with Bonferroni post hoc; data are represented as mean ± SEM from 3 independent experiments (n = 3). *P < 0.05 between the indicated groups.