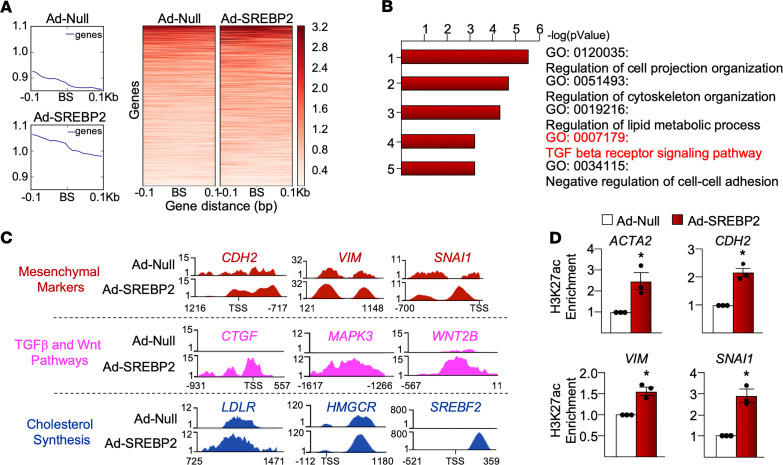
Figure 3. ATAC-Seq reveals SREBP2 modulation of the chromatin accessibility.
HUVECs were infected with Ad-SREBP2(N) or empty vector (Ad-Null) followed by ATAC-Seq (n = 2). (A) ATAC peaks were merged into a union set. SREBP2 putative binding sites within the ATAC peaks were predicted. The upper and lower panels show overall and per-loci ATAC signals at ± 0.1 kb flanking the putative SREBP2 binding sites (BS), respectively. (B) Genes specific to mesenchymal markers and TGF-β/Wnt pathways indicate chromatin decondensation upon SREBP2(N) overexpression by using WashU Epigenome Browser. Cholesterol biosynthesis genes were used as a positive control. (C) Gene ontology (GO) analysis reveals top activated pathways with SREBP2(N) transactivation. (D) H3K27ac ChIP-qPCR indicates the chromatin remodeling of the mesenchymal genes. Data in D were analyzed by 2-tailed Student’s t test; data are represented as mean ± SEM from 3 independent experiments (n = 3). *P < 0.05 between the indicated groups. ACTA2, α smooth muscle actin; CDH2, neural cadherin; CTGF, connective tissue growth factor; HMGCR, 3-Hydroxy-3-Methylglutaryl-CoA Reductase; LDLR, low density lipoprotein receptor; MAPK3, mitogen activated protein kinase 3; SNAI1, snail family transcriptional repressor 1; TGF, transforming growth factor; VIM, vimentin; Wnt, wingless integration site.