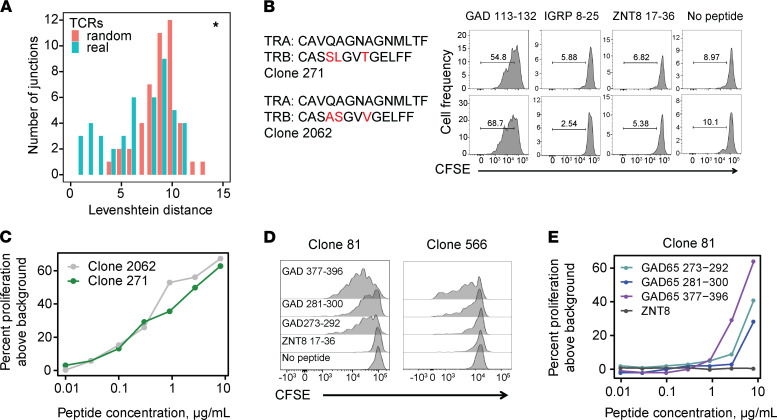
Figure 5. TRB junctions sharing a common public TRA junction differ in sequence but may have similar roles in binding.
(A) TRB junctions that share TRA junctions show greater sequence identity than expected by chance. Numbers of TRB junctions paired with unique public and private TRA junctions were calculated from n = 72 and 165 public and private TRA junctions, respectively (Supplemental Table 3, public/private TCRs). Levenshtein distances were calculated for paired combinations of unique TRB junctions that pair with public TRA junctions (n = 31). For null sets, Levenshtein distances were calculated for TRB junctions in equal-sized, random sets of nonexpanded TRB junctions (n = 31 junctions) (Supplemental Table 3), and this was repeated n = 1000 times. Shown is a histogram representative of the median difference between real and random sets, as judged by P values from Kolmogorov-Smirnov tests. *, P value < 0.05. (B) TCR clones sharing public TRA chains with mismatched TRB junctions were functionally triggered by the same peptides. Recombinant TCR clones (Clone_271 and Clone_2062, Table 2) were transduced into primary CD4+ T cells, and proliferation was measured using a dye dilution assay following stimulation with the indicated peptides or a no-peptide control. Red font, mismatched residues. (C) Clone_271 and Clone_2062 TCR clones share similar dose-response curves for the GAD 113–132 peptide. (D) Cross-reactivity of related TCR clones, Clone_81 and Clone_566, for multiple GAD65 peptides. GAD 273–292 and GAD281–300 have overlapping sequences, but GAD 377–396 is distinct (Supplemental Table 2). (E) Dose-response curves showing cross-reactivity of Clone_81 for multiple GAD65 peptides and a nonreactive ZNT8 peptide.