Abstract
Japanese encephalitis (JE) is one of the viral diseases affecting millions of peoples across the globe specifically developing countries. There is no specific treatment available, however, vaccines are available for its prevention. Unfortunately, available vaccines are not effective against all clinical isolates and are also associated with neurological complications in some individuals. We have screened the selected phytoconstituents of Andrographis paniculata against various targets of Japanese encephalitis virus (JEV) using Schrodinger suite 2019-3. Among all selected phytoconstituents, andrographolide has shown a good binding affinity towards NS3 protease as compared to NS3 helicase and NS5 Rdrp (RNA dependent RNA polymerase) of JEV. The molecular dynamics (MD) results have also shown good stability of andrographolide in the active site of NS3 protease. The absorption, distribution, metabolism, excretion, and toxicity (ADMET) analysis has also indicated a good pharmacokinetic and safety profile of andrographolide. Finally, the in-vitro target-based assay have confirmed the inhibitory potential of andrographolide against the NS3 protease of JEV. In conclusion, andrographolide could have the potential to develop as an antiviral agent against JEV through inhibition of protease, however, further investigations are required.
Keywords: Andrographis paniculata, Japanese encephalitis virus, Computational studies, In-vitro study, NS3 protease
Graphical abstract

Highlights
-
•
Andrographolide has shown stable binding conformation in the active site of protease of JEV.
-
•
The protease of JEV was inhibited in a concentration dependent manner.
1. Introduction
Japanese encephalitis (JE) is mainly undesired inflammation of the brain which is caused by the Japanese encephalitis virus (JEV), one of the Flavivirus belonging to the family Flaviviridae. There are more than 3 billion peoples at risk of JEV infection across 24 countries. In Asia, approximately 68,000 clinical viral encephalitis cases are found every year (WHO, 2021). In India, the most affected state is Uttar Pradesh where around 1000 children are died due to JE. It has been observed that JEV easily crosses the blood-brain-barrier (BBB) and results in neurological disorders (King et al., 2007; Mathur et al., 1988). The JEV is a positive sense RNA (ribonucleic acid) virus that contains an open reading frame (ORF) that codes for polyprotein which further slashed by viral and host proteases into 10 proteins out of which 7 are non-structural (NS) proteins named NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 and remaining 3 are structural proteins named Capsid (C), pre-membrane (PrM), and envelope (E) (Bhimaneni and Kumar, 2020; Navyashree et al., 2021). Currently, there is no specific treatment for JE (WHO, 2021), only symptomatic treatments are available. Vaccination has been used for JE but neurological complications, allergic reactions limit their use (Schuller et al., 2011; Plesner, 2003).
Herbal products have shown promising effects against JEV (Ganjhu et al., 2015), however, as we know that standardization, stability, uniformity, etc. are major concerns associated with herbal products. Thus, there is a need for alternative approaches. Andrographis paniculata has been evaluated for its antiviral activities. The antiviral potential of ethanolic extract of Andrographis paniculata has been reported against Dengue virus belonging to genus flavivirus, family Flaviviridae (Ramalingam et al., 2018). However, the extract role of phytoconstituents of Andrographis paniculata against JEV is still unclear. Thus, the aim of the present study was to screen the selected phytoconstituents of Andrographis paniculata against targets of JEV using molecular docking, molecular dynamics (MD) approach followed by target specific NS3 protease enzymatic assay.
2. Material and methods
2.1. Chemicals
Andrographolide was purchased from TCI Chemicals (India) Pvt. Ltd. Fluorescent substrate Boc-Gly-Lys-Arg-MCA (5 mg vial) MGK-3143-v was purchased from Peptide Institute. The Recombinant West Nile Virus NS3 Protease Protein was purchased from Biotech India Pvt. Ltd. All other chemicals are purchased from a suitable vendor.
2.2. In-silico study
2.2.1. Protein structure and ligands preparation
The crystal structures of JEV target proteins i.e., NS3 helicase ((PDB ID: 2Z83), NS5 (PDB ID: 4K6M), and NS3 protease (PDB ID: 4R8T) were downloaded from the Research Collaboratory for Structural Bioinformatics, Protein Data Bank (RCSB PDB) (Rose et al., 2016). Proteins were processed, optimized, and minimized in the protein preparation wizard. Formerly, all hydrogen atoms were added to these specified JEV protein structures. The geometry of these protein targets is corrected using default parameters such as PH ionization, energy minimization. The phytoconstituents of Andrographis paniculata were retrieved through literature and their corresponding structures were obtained from PubChem (Fig. S1). These obtained structures were drawn with the help of Chem-Bio-Draw 14.0 (PerkinElmer Informatics, Waltham, MA, USA). These structures were prepared using Lig Prep tool of Schrödinger Suite 2019-3. Various possible isomers were generated for these ligands out of which, the best one is selected for docking against the prepared targets (Banks et al., 2005).
2.2.2. Docking study
Docking study was conducted in the extra precision (XP) mode of the Schrödinger 2019-3 suite. The target protein receptor grid was generated by using the Maestro tool of Schrödinger suite 2019-3. The grid box was generated based on the presence of an internal ligand in that particular receptor (Friesner et al., 2006). The active site of NS5 (4K6M) was generated using an internal ligand whereas site mapping was done for NS3 helicase (2Z83) and NS3 protease (4R8T). The prepared phytoconstituents were docked against specified protein targets of JEV such as NS3 helicase ((PDB ID: 2Z83), NS5 (PDB ID: 4K6M), and NS3 protease (PDB ID: 4R8T). The OPLS3e force field was used and possible stable configuration of ligands in the active site of targets is selected based on doc scores. The interactions between the ligand and protein were characterized through the formation of hydrogen bonding, π- π, van der Waals, and hydrophobic interactions (Friesner et al., 2006; Gálvez et al., 2018).
2.2.3. ADMET profile
The ADMET profile of andrographolide was predicted using the pkCSM online web server available at http://biosig.unimelb.edu.au/pkcsm/prediction# (Pires et al., 2015). The drug likeliness was predicted using Molinspiration online web server based on “Lipinski's Rule of Five” (Jarrahpour et al., 2012; Lipinski et al., 2012). The LD50 value was predicted using the ProTox webserver (Banerjee et al., 2018).
2.2.4. Molecular dynamics (MD) studies
The time-dependent behavior of the ligand protein complex was analyzed using molecular dynamics studies (Blessy et al., 2015; Oliveira et al., 2019). MD studies were carried out using the Desmond module of Schrödinger 2019-3 suite. A simple point charge (SPC) model was used to incorporate protein-ligand complex into an orthorhombic water box. The orthorhombic box generates a 10 Å buffer region between the protein atoms and box. This system was further solvated by the TIP3P water model (Blessy et al., 2015). The water molecules were removed and the system was neutralized by the addition of Na+ ions. The energy calculation was done using the OPLS3e force field. The temperature and pressure were maintained at a constant 300 K and 1.01325 bar respectively. The model was relaxed before giving simulation for 10 ns. The root mean square deviation (RMSD) and root mean square fluctuation (RMSF) values were calculated.
2.3. In-vitro target-based study
2.3.1. Protease inhibitory assay
The protease inhibitory assay was performed (Fig. S2) as mentioned in vendor protocol. Briefly, the protease was reconstituted in deionized water as 1 ng/μL and kept constant throughout the assay. The fluorescent substrate of concentration 40 μM was prepared in distilled water. Initially, a volume of 20 μL of TRIS buffer was added in the Tarson 96 well plate followed by the addition of 10 μL of protease (1 ng/μL). Further, a volume of 10 μL of andrographolide at different concentrations (250 μg/ml-1.95 μg/ml) was added and incubation was carried out with viral NS3 protease at 25 °C for 15 min. Finally, 10 μL of the fluorescent substrate was added and final incubation was done for 15 min at 25 °C (Oliveira et al., 2019). The fluorescence (excitation: 380 nm, emission: 460 nm) was measured by using the BioTek Synergy H1 microplate reader.
3. Results
3.1. Computational studies
The particular docking scores were obtained upon the binding of phytoconstituents to the specific JEV targets. The lower docking score indicates the high and optimum binding affinity of constituents towards the corresponding target. Molecular docking is widely used to predict energetically favorable binding conformations of ligands within the active sites of the target protein (Gupta et al., 2018). This binding was accompanied with formation of hydrogen bonds, hydrophobic interactions such as Pi-Pi, Pi-cation, etc. These all above interactions indicate the stability of complexes which is formed after the binding of ligand to target protein. There are around 30 phytoconstituents present in Andrographis paniculata (Tan et al., 2016). All 30 phytoconstituents of Andrographis paniculata have been screened against various targets of JEV. The doc score of individual phytoconstituents of Andrographis paniculata against NS3 helicase, NS5 (RdRp) and NS3 protease has been compiled in table S1, S2 and S3 respectively (supplementary file). Among these phytoconstituents, andrographolide (Fig. 1) has shown optimum binding affinity with NS3 helicase, NS3 protease and NS5 of JEV which is described below.
Fig. 1.
Structure of andrographolide.
3.1.1. Interaction of andrographolide with NS3 helicase of JEV
Andrographolide has shown an optimum binding affinity towards NS3 helicase of JEV through hydrogen bonding and hydrophobic interactions. The hydrogen bonding was observed with VAL 430, HIS 488, THR 290, and SER 412 whereas hydrophobic interaction was observed with PHE 289, ALA 453, ILE 411, VAL 430, PRO 432, and LEU 444 of active site amino acids of NS3 helicase of JEV as shown in Fig. 2. The docking score of andrographolide was found to be −5.7 kcal/mol whereas the doc score of reference i.e., paritaprevir was found to be −3.86 kcal/mol as shown in Table 1.
Fig. 2.
Interactions of andrographolide towards active site of NS3 helicase of JEV using Schrödinger 2019-3 suite.
Table 1.
Interactions of andrographolide against NS3 Helicase, NS5 and NS3 Protease along with their binding affinity and contacting amino acids residues.
| Phytoconstituent Name | Binding Affinity (Kcal/M) | Residues interacting with Ligand through H-Bonding | Closer Contact Residues |
|---|---|---|---|
| A. NS3 Helicase | |||
| Andrographolide | −5.7 | Val430, His488, Thr 290, Ser412 | Asp410, Ile411, Gln457, Ser454, Ala453, Thr451, Phe289, Ser429, Lys431, Leu444, Arg600 |
| Reference (Paritaprevir) | −3.8 | Glu155 | Leu49, Lys73, Glu74, Asp75, Trp83, Phe85, Asp86, Arg87, Lys88, Thr118, Leu120, Gly121, Glu122, Val13, Ala125, Ile147, Gly148, Leu149, Gly151, Asn152, Gly153, Val154, Leu156 |
| B. NS5 | |||
| Andrographolide | −4.5 | Asp146 | Glu111, Gly81, Cys82, Gly83, Arg84, Trp87, Ile147, Gly148, Ser150, Tyr220, Glu218, Asn216, Arg213 |
| Reference (S-Adenosyl-L-Homocysteine) | −10.112 | Ser46, Gly86, Asp131, Val132, Asp146 | Val55, Gly58, Trp87, Gly81, Cys82, Gly83, Gly85, Thr104, Hid110, Val130, Phe133, Ile147, Gly148 and Glu149 |
| C. NS3 Protease | |||
| Andrographolide | −6.6 | Lys73, Asn152 | Arg76, Glu74, Lys73, Leu120, Val154, Gly153 |
| Reference (ST-610) | −5.5 | Thr290 and Asp410 | Phe289, Asp291, Ala364, Thr409, Ser412, Ser429, Val430, Lys431, Pro432, Thr451, Ala453, Ser454, Gln457, Arg458 and His488 |
3.1.2. Interactions of andrographolide with NS5 of JEV
Andrographolide has interacted through hydrogen and hydrophobic interactions with NS5 of JEV as shown in Fig. 3. The docking score was found to be −4.5 kcal/mol whereas the doc score of internal ligands i.e., S-Adenosyl-L-Homocysteine was found to be −10.11 kcal/mol as shown in Table 1.
Fig. 3.
Interactions of andrographolide towards active site of NS5 of JEV using Schrödinger 2019-3 suite.
3.1.3. Interactions of andrographolide with NS3 protease of JEV
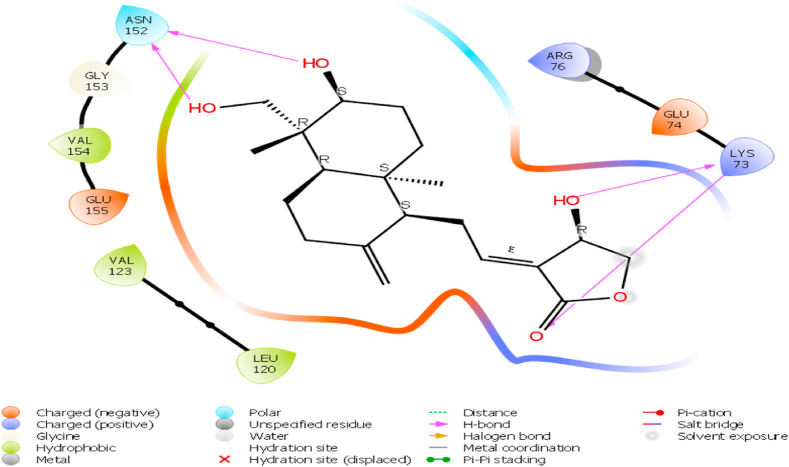
Andrographolide has also shown optimum binding affinity against NS3 protease of JEV through hydrogen bonding with LYS 73, and ASN 152 as shown in Fig. 4. The doc score of andrographolide was found to be −6.6 kcal/mol whereas doc score of internal ligands i.e., ST-610 was found to be −5.5 Kcal/Mole as shown in Table 1.
Fig. 4.
Interactions of andrographolide towards active site of NS3 protease of JEV using Schrödinger 2019-3 suite.
3.1.4. ADMET prediction
The andrographolide follows Lipinski's rule of five which indicates drug likeliness characteristics. The Caco-2 permeability, gastro intestinal absorption (GIT) absorption, AMES toxicity, Intestinal absorption falls under the acceptable range as shown in Table 2. The predicted LD50 value of andrographolide was found to be 5000 mg/kg which indicates a good safety profile as shown in Table 2.
Table 2.
ADMET profile of Andrographolide.
| Models | Andrographolide |
|---|---|
| Molecular weight (g/mol) | 336.42 |
| Log P O/W | 2.55 |
| GI absorption | High |
| BBB permeability | No |
| Caco-2 Permeability (log Papp in 10−6 cm/s) | 1.07 |
| Intestinal absorption (% Absorbed) | 95.357 |
| P-glycoprotein Substrate | No |
| P-glycoprotein 1 Inhibitor | No |
| P-glycoprotein 2 Inhibitor | No |
| VDss (human) (log L/kg) | −0.286 |
| BBB permeability (log BB) | −0.598 |
| CNS permeability (log PS) | −2.691 |
| C. Metabolism | |
| CYP2D6 substrate | No |
| CYP3A4 substrate | Yes |
| CYP1A2 inhibitor | No |
| CYP2C19 inhibitor | No |
| CYP2C9 inhibitor | No |
| D. Excretion | |
| Total Clearance (log ml/min/kg) | 1.183 |
| Renal OCT2 substrate | No |
| E. Toxicity | |
| AMES toxicity | No |
| Max. tolerated dose (human) (log mg/kg/day) | 0.128 |
| hERG I inhibitor | No |
| hERG II inhibitor | No |
| Hepatotoxicity | No |
| Skin Sensitisation | No |
| T. Pyriformis toxicity (log mM) | 0.491 |
| F. LD50(mg/kg) | 5000 mg/kg |
Caco-2 Permeability: >0.90 (High); Intestinal absorption: <30% (Poor absorption); Volume of distribution: >0.48 (High) & <0.15 (Low); Intestinal absorption: <30 (Poorly absorbed); Skin Permeability: >-2.5 (low); BBB permeability: >0.3 (crosses BBB), <-1 (poor); CNS permeability: >-2 (penetrable to CNS), <-3 (unable to penetrate CNS); T. Pyriformis toxicity (log mM): >-0.5 (Toxic).
3.1.5. Molecular dynamics (MD) study
Molecular docking results have indicated a good binding affinity of andrographolide with NS3 protease as compared to NS3 helicase and NS5 protease of JEV. Thus, further MD study was conducted to check the stability of andrographolide in the active site of NS3 protease of JEV. The results of molecular dynamics are mentioned below.
3.1.5.1. Root mean square deviation (RMSD) analysis
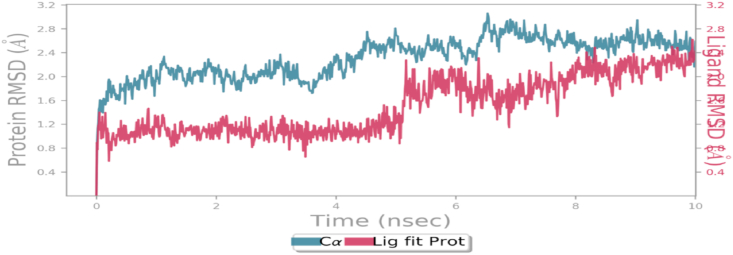
Fig. 5 represents an analysis of RMSD of andrographolide and NS3 protease of JEV. The results have shown instability of ligand initially, however after 8 ns it becomes stable as shown in Fig. 5.
Fig. 5.
Root Mean Square Deviation (RMSD) of backbone skeleton of protein NS3 protease and ligand as andrographolide using Desmond module of Schrödinger 2019-3 suite.
3.1.5.2. Root mean square fluctuation (RMSF) and protein secondary structure analysis
RMSF is useful to determine the flexibility of proteins on the basis of fluctuation of amino acid residues. It was found that the RMSF value of NS3 protease of JEV was similar to that of protein with ligand complex. The observed peaks were the indication of fluctuation of specific region of protease of JEV. The amino acid residues of specific positions were not fluctuating with respect to time. The N- and C- terminal of the protein fluctuated more as compared to the other parts of the protein. The secondary structures of protein such as alpha helixes and beta plated sheets were enough rigid to stay in stationary position without fluctuating as shown in Fig. 6.
Fig. 6.
RMSF of andrographolide upon binding with the protein using Desmond module of Schrödinger 2019-3 suite.
The alpha helices and beta sheets of proteins were analyzed throughout the simulation by using Schrodinger suite 2019-3. Fig. 7 represents each residue and its secondary structure elements (SSE) with respect to the simulation.
Fig. 7.
Effect of molecular dynamics simulation on secondary structure elements (SSE) using Desmond module of Schrödinger 2019-3 suite.
3.1.5.3. Protein-ligand contacts analysis
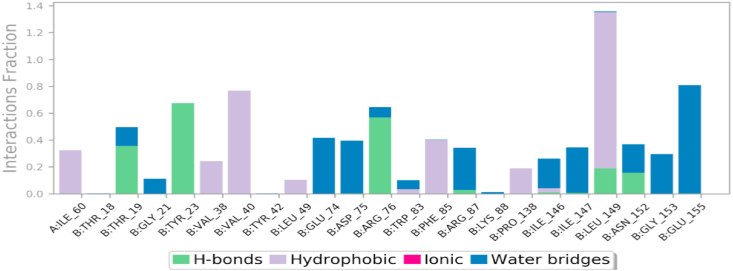
The andrographolide and NS3 protease of JEV contact analysis was shown in Fig. 8. The hydrophobic interactions are predominant as compared to other interactions. The highest hydrogen bond interaction occurred with leucine 149 present in the B chain (Fig. 8).
Fig. 8.
Histogram indicating the interaction of andrographolide with NS3 protease in terms of interaction fraction using Desmond module of Schrödinger 2019-3 suite.
3.2. In-vitro target-based assay
The inhibitory activity of andrographolide against NS3 protease of JEV was determined. The results have shown inhibition of NS3 protease of JEV by andrographolide in concentration-dependent manner. The IC50 value of andrographolide was found to be 2 μg/ml as shown in Fig. S3.
4. Discussion
The current investigation has screened selected phytoconstituents of Andrographis paniculata against various targets of JEV. Literature has shown various biological activities such as anti-inflammatory, anti-angiogenesis, antiproliferation, neuroprotective and hepatoprotective effects of Andrographis paniculata (Chan et al., 2010; Maiti et al., 2010). The antiviral properties are also reported against human immunodeficiency virus (Uttekar et al., 2012), herpes simplex virus 1 (Seubsasana et al., 2011), influenza virus (Chan et al., 2010), and Epstein–Barr virus (Lin et al., 2008). The antiviral activity of andrographolide one of the phytoconstituent of Andrographis paniculata against the Dengue and Hepatitis C virus is also reported (Panraksa et al., 2017; Lee and Yun, 2014). However, to the best of our knowledge, no study is reported so far regarding the antiviral activity of Andrographis paniculata against JEV. Currently, no specific drug is available in the treatment of JE. Thus, we have screened phytoconstituents of Andrographis paniculata against various targets of JEV.
Molecular docking is one of the well-known computational techniques used to predict the energetically favored configuration of ligand in the active site of the target (Vyas et al., 2008). Japanese encephalitis virus contains structural (capsid, envelope, and PrM protein) and non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) that are formed after cleavage of polyprotein with the help of protease (Geiss et al., 2009). Among all selected phytoconstituents, andrographolide has shown energetically favored configuration in the active site of protease of JEV. Another nonstructural protein NS3 helicase also plays an important role in the separation of dsRNA that is formed at the time of viral replication (Gallivan et al., 2003). The proteins that are attached to the viral RNA during replication are mainly removed with the help of NS3 helicase (Malet et al., 2008). The results of current investigation have shown an optimum binding affinity of andrographolide towards NS3 helicase protein. The RNA dependent RNA polymerase (RdRp) acts as one of the prominent targets in the development of antiviral drugs due to its tendency of synthesizing both negative and positive-strand genomic RNA (Malet et al., 2008). The flavivirus NS5 N-terminal contains methyltransferase (MTase) whereas C-terminal contains RNA-dependent RNA polymerase (RdRP) (Geiss et al., 2009). The results of current investigation have also shown good binding affinity towards NS5 of JEV. Overall, andrographolide has shown a good binding affinity towards NS3 protease of JEV as compared to NS3 helicase and NS5 Rdrp of JEV. Thus, further molecular dynamics studies were conducted with NS3 protease of JEV. The results of molecular dynamics have shown stable configuration of andrographolide with NS3 protease of JEV.
In drug discovery and development, the absorption profile of a compound plays an integral role. The ADMET inspection of compounds helps to distinguish compounds with poor absorption profiles which will be beneficial in the early stage of drug development (Kant et al., 2019). The ADMET profile of andrographolide was analyzed using Pro Tox, PkCSM, and Molinspiration online web server. The drug likeliness is one of the important aspects of compounds that are in the drug discovery and development phase which was predicted by the Lipinski rule of five. It has been found that andrographolide follows the Lipinski rule of five which supports its drug-like properties.
The inhibitory potential of andrographolide against protease of JEV was confirmed through experimental study. The results have shown inhibition of JEV NS3 protease which helps in cleavage of polyprotein of JEV into the structural and nonstructural viral proteins by andrographolide in a concentration dependent manner. Thus, andrographolide could be developed as protease inhibitor against JE. The possible mechanism of andrographolide as anti-JEV agent is explained in Fig. S4.
5. Conclusion
Andrographolide has shown an optimum binding affinity towards the protease as compared to NS3 helicase and NS5 Rdrp of JEV. The molecular dynamics results have confirmed the stability of andrographolide in the active site of protease of JEV. The in-vitro target-based assay has confirmed the inhibitory potential of andrographolide against protease of JEV. Overall, it can be concluded that andrographolide has the potential to develop as an effective anti-JEV agent. However, further, specific antiviral assays such as cell viability, plaque reduction, immunofluorescence assay, etc. are required to confirm its antiviral activity against JEV.
CRediT authorship contribution statement
Shailesh Bhosale: Investigation, Data curation, Software, Writing – original draft. Anoop Kumar: Conceptualization, Methodology, Supervision, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are thankful to S.J.S. Flora, Director, National Institute of Pharmaceutical Education and Research (NIPER), Raebareli, Lucknow (UP)-India for providing the necessary facility to carry out this research work. Author, Anoop Kumar is also thankful to Prof. R.K. Goyal, Vice Chancellor, Delhi Pharmaceutical Sciences and Research University (DPSRU)-New Delhi for continuous support and motivation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crphar.2021.100043.
Compliance with ethical standards
Not applicable.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46(W1):W257–W263. doi: 10.1093/nar/gky318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks J.L., Beard H.S., Cao Y., et al. Integrated modeling program, applied chemical theory (IMPACT) J. Comput. Chem. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhimaneni S.P., Kumar A. Abscisic acid, a plant hormone, could be a promising candidate as an anti-Japanese encephalitis virus (JEV) agent. Anti-Infective Agents. 2020;18(4) doi: 10.2174/2211352518666200108092127. [DOI] [Google Scholar]
- Blessy J.J., Sharmila D.J.S. Molecular simulation of N-acetylneuraminic acid analogs and molecular dynamics studies of cholera toxin-Neu5Gc complex. J. Biomol. Struct. Dyn. 2015;33(5):1126–1139. doi: 10.1080/07391102.2014.931825. [DOI] [PubMed] [Google Scholar]
- Chan S.J., Wong W.F., Wong P.T., Bian J.S. Neuroprotective effects of andrographolide in a rat model of permanent cerebral ischaemia. Br. J. Pharmacol. 2010;161:668–679. doi: 10.1111/j.1476-5381.2010.00906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Murphy R.B., Repasky M.P., et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein− ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Gallivan J.P., McGarvey M.J. The importance of the Q motif in the ATPase activity of a viral helicase. FEBS Lett. 2003;554:485–488. doi: 10.1016/s0014-5793(03)01229-8. [DOI] [PubMed] [Google Scholar]
- Gálvez J., Polo S., Insuasty B., Gutiérrez M., et al. Design, facile synthesis, evaluation of novel spiro- pyrazolo [1, 5-c] quinazolines as cholinesterase inhibitors: molecular docking MM/GBSA studies. Comput. Biol. Chem. 2018;74:218–229. doi: 10.1016/j.compbiolchem.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Ganjhu R.K., Mudgal P.P., Maity H., Dowarha D., Devadiga S., Nag S., Kumar A.G. Herbal plants and plant preparations as remedial approach for viral diseases. Virusdisease. 2015;26(4):225–236. doi: 10.1007/s13337-015-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss B.J., Stahla H., Hannah A.M., Gari H.H., Keenan S.M. Focus on flaviviruses: current and future drug targets. Future Med. Chem. 2009;1:327–344. doi: 10.4155/fmc.09.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Kant K., Sharma R., Kumar A. Evaluation of in silico anti-Parkinson potential of β-asarone. Cent. Nerv. Syst. Agents Med. Chem. 2018;18(2):128–135. doi: 10.2174/1871524918666180416153742. [DOI] [PubMed] [Google Scholar]
- Jarrahpour A., et al. Osiris and Molinspiration (POM) together as a successful support in drug design: antibacterial activity and biopharmaceutical characterization of some azo Schiff bases. Med. Chem. Res. 2012;21:1984–1990. [Google Scholar]
- Kant K., Lal U.R., Kumar A., Ghosh M. A merged molecular docking, ADME-T and dynamics approaches towards the genus of Arisaema as herpes simplex virus type 1 and type 2 inhibitors. Comput. Biol. Chem. 2019;78:217–226. doi: 10.1016/j.compbiolchem.2018.12.005. [DOI] [PubMed] [Google Scholar]
- King N.J., Getts D.R., Getts M.T., Rana S., Shrestha B., Kesson A.M. Immunopathology of flavivirus infections. Immunol. Cell Biol. 2007;85:33–42. doi: 10.1038/sj.icb.7100012. [DOI] [PubMed] [Google Scholar]
- Lee Y.M., Yun S.I. Japanese encephalitis: the virus and vaccines. Hum. Vaccines Immunother. 2014;10(2):263–279. doi: 10.4161/hv.26902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.P., Chen S.Y., Duh P.D., Chang L.K., Liu Y.N. Inhibition of the epstein-barr virus lytic cycle by andrographolide. Biol. Pharm. Bull. 2008;31(11):2018–2023. doi: 10.1248/bpb.31.2018. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Maiti K., Mukherjee K., Murugan V., Saha B.P., Mukherjee P.K. Enhancing bioavailability and hepatoprotective activity of andrographolide from Andrographis paniculata, a well-known medicinal food, through its herbosome. J. Sci. Food Agric. 2010;90:43–51. doi: 10.1002/jsfa.3777. [DOI] [PubMed] [Google Scholar]
- Malet H., Massé N., Selisko B., Romette J.L., Alvarez K., Guillemot J.C., Tolou H., Yap T.L., Vasudevan S.G., Lescar J., Canard B. The flavivirus polymerase as a target for drug discovery. Antivir. Res. 2008;80:23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Mathur A., Bharadwaj M., Kulshreshtha R., Rawat S., Jain A., Chaturvedi U.C. Immunopathological study of spleen during Japanese encephalitis virus infection in mice. Br. J. Exp. Pathol. 1988;69:423–432. [PMC free article] [PubMed] [Google Scholar]
- Navyashree V., Kant K., Kumar A. Natural chemical entities from Arisaema Genus might be a promising break-through against Japanese encephalitis virus infection: a molecular docking and dynamics approach. J. Biomol. Struct. Dyn. 2021;39:1404–1416. doi: 10.1080/07391102.2020.1731603. [DOI] [PubMed] [Google Scholar]
- Oliveira A.S., Gazolla P.A., Oliveira A.F.C.D.S., Pereira W.L., Viol L.C.D.S., Maia A.F.D.S., Santos E.G., da Silva Í.E., de Oliveira Mendes T.A., da Silva A.M., Dias R.S. Discovery of novel West Nile Virus protease inhibitor based on isobenzonafuranone and triazolic derivatives of eugenol and indan-1, 3-dione scaffolds. PloS One. 2019;14(9) doi: 10.1371/journal.pone.0223017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panraksa P., Ramphan S., Khongwichit S., Smith D.R. Activity of andrographolide against dengue virus. Antivir. Res. 2017;139:69–78. doi: 10.1016/j.antiviral.2016.12.014. [DOI] [PubMed] [Google Scholar]
- Pires D.E., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesner A.M. Allergic reactions to Japanese encephalitis vaccine. Immunol. Allergy Clin. 2003;23:665–697. doi: 10.1016/s0889-8561(03)00102-4. [DOI] [PubMed] [Google Scholar]
- Ramalingam S., Karupannan S., Padmanaban P., Vijayan S., Sheriff K., Palani G., Krishnasamy K.K. Anti-dengue activity of Andrographis paniculata extracts and quantification of dengue viral inhibition by SYBR green reverse transcription polymerase chain reaction. Ayu. 2018;39:87–91. doi: 10.4103/ayu.AYU_144_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z., Green R.K. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2016;45(D1):D271–D281. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller E., Klingler A., Dubischar-Kastner K., Dewasthaly S., Müller Z. Safety profile of the Vero cell-derived Japanese encephalitis virus (JEV) vaccine IXIARO®. Vaccine. 2011;29(47):8669–8676. doi: 10.1016/j.vaccine.2011.08.117. [DOI] [PubMed] [Google Scholar]
- Seubsasana S., Pientong C., Ekalaksananan T., Thongchai S., Aromdee C. A potential andrographolide analogue against the replication of herpes simplex virus type 1 in vero cells. Med. Chem. 2011;7:237–244. doi: 10.2174/157340611795564268. [DOI] [PubMed] [Google Scholar]
- Tan M.C., Oyong G.G., Shen C.C., Ragasa C.Y. Chemical constituents of Andrographis paniculata (Burm. f.) nees. Int J Pharmacogn Phytochem Res. 2016;8(8):1398–1402. [Google Scholar]
- Uttekar M.M., Das T., Pawar R.S., Bhandari B., Menon V., Gupta S.K., Bhat S.V. Anti-HIV activity of semisynthetic derivatives of andrographolide and computational study of HIV-1 gp120 protein binding. Eur. J. Med. Chem. 2012;56:368–374. doi: 10.1016/j.ejmech.2012.07.030. [DOI] [PubMed] [Google Scholar]
- Vyas V., Jain A., Jain A., Gupta A. Virtual screening: a fast tool for drug design. Sci. Pharm. 2008;76:333–360. [Google Scholar]
- World Health Organization (WHO) Fact Sheets on Japanese encephalitis. Available at https://www.who.int/news-room/fact-sheets/detail/japanese-encephalitis. Accessed on 14th April 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.