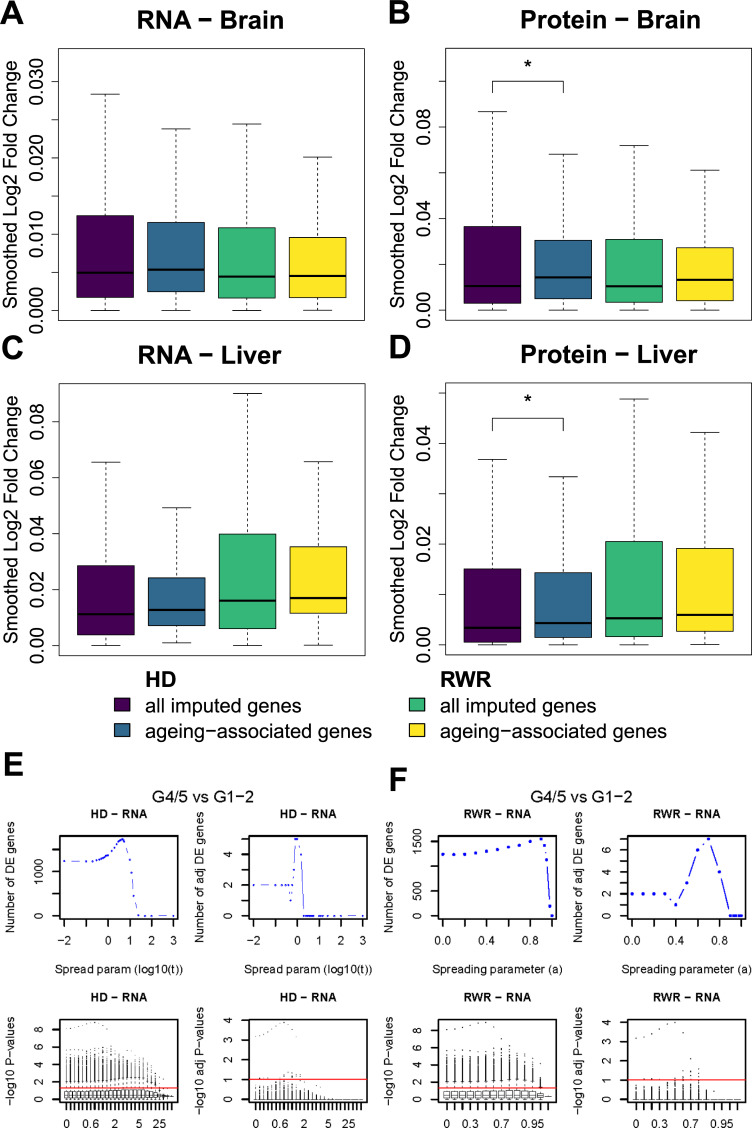
Fig 6. Network propagation identifies ageing-associated genes as well as genes distinguishing prostate tumors of different grades.
RNA (A and C) and proteome (B and D) log2 fold changes were smoothed on the network using RWR (yellow/green colored) and HD (purple/blue colored). The spreading parameters were set to α = 0.5 and t = 0.7 for brain (A and B) and α = 0.4 and t = 0.3 for liver (C and D). These are the optimal parameters found by the between-dataset consistency analysis. Afterwards, we subset unmeasured genes which were recovered using network propagation and identified ageing-associated genes within these imputed genes. The median absolute propagated log2 fold change of the ageing-associated genes within imputed genes is higher than the median absolute propagated log2 fold change of all imputed genes. In the case of protein log2 fold changes in brain and liver smoothed with HD the difference is significant (one-sided Wilcoxon rank sum test; * p ≤ 0.05). E: Number of differentially expressed genes between the more and less aggressive PCa tumors with HD at the mRNA layer before (upper left) and after multiple hypothesis testing correction (upper right) across the values of the spreading parameter. Below, distributions of the negative logarithm in base 10 of the t-test p-values for the different values of the spreading parameter before (lower left) and after multiple hypothesis testing correction (lower right) are depicted. The red lines have ordinate -log10(0.05) and -log10(0.1) respectively and correspond to the significance threshold. F: Same as in (E) but for RWR.