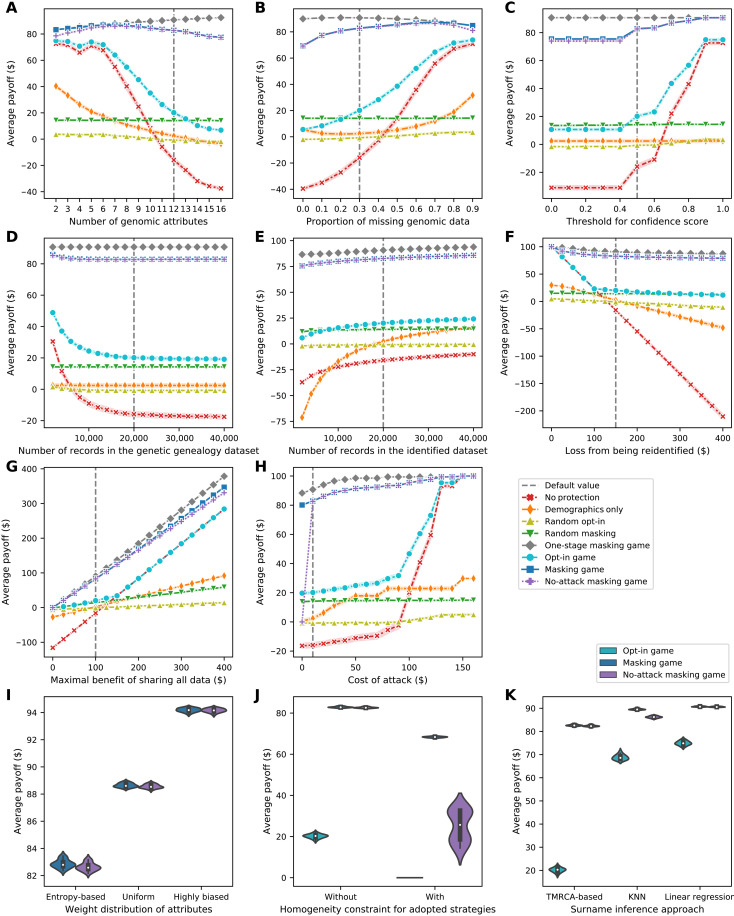
Fig. 4. Sensitivity of the data subjects’ average payoff as a function of the parameters and settings in the model.
(A) Line plot of the sensitivity to the number of genomic attributes. (B) Line plot of the sensitivity to the proportion of missing genomic data. (C) Line plot of the sensitivity to the threshold for confidence score. (D) Line plot of the sensitivity to the number of records in the genetic genealogy dataset. (E) Line plot of the sensitivity to the number of records in the identified dataset. (F) Line plot of the sensitivity to the loss from being re-identified. (G) Line plot of the sensitivity to the maximal benefit of sharing all data. (H) Line plot of the sensitivity to the cost of an attack. (I) Violin plot of payoff distribution’s sensitivity to the strategy adoption setting. (J) Violin plot of the sensitivity to the surname inference approach. (K) Violin plot of the sensitivity to the weight distribution of attributes. Each line plot (depicted using Seaborn) shows data subjects’ average payoffs, with error bars representing SDs, in eight scenarios. Each violin plot (depicted using Seaborn) combines boxplot and kernel density estimate for showing the distributions of data subjects’ average payoffs in several scenarios. Gaussian kernels are used with default parameter settings. TMRCA, time to most recent common ancestor; KNN, k-nearest neighbors.