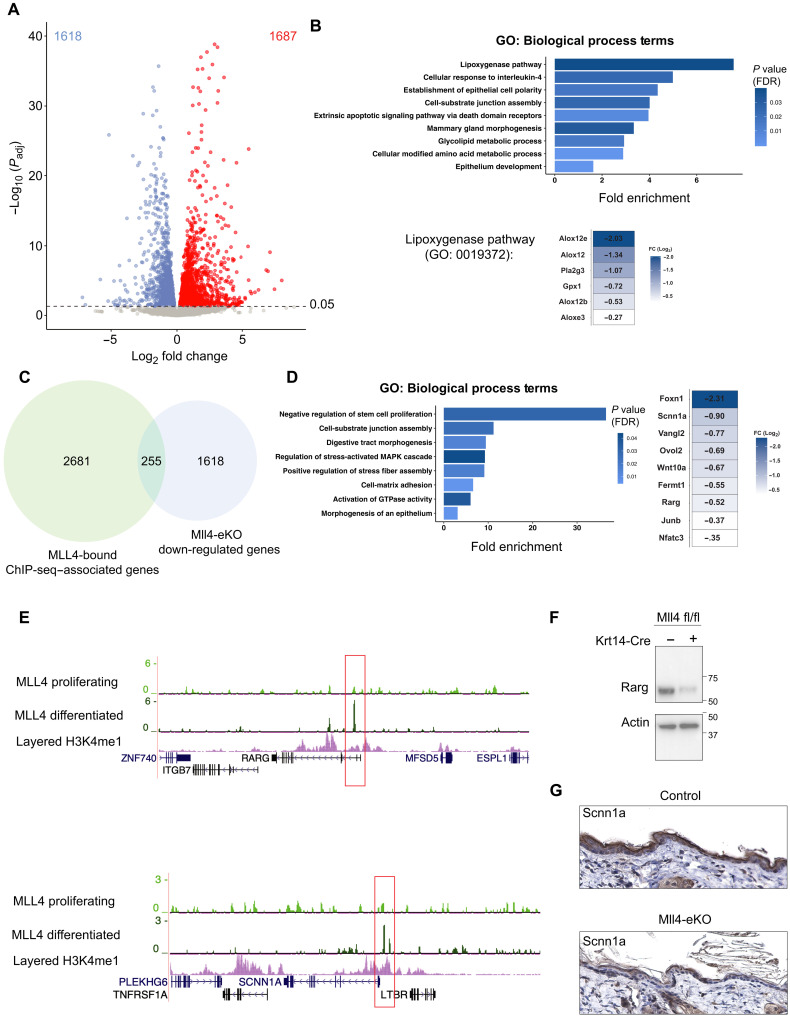
Fig. 2. Loss of Mll4 leads to a profoundly altered transcriptome characterized by a loss of key genes involved in differentiation and lipid metabolism.
(A) Differentially expressed genes (1618 down-regulated genes and 1687 up-regulated genes) in isolated bulk epidermis of 3-week-old Mll4-eKO and control mice. Dashed line denotes adjusted P value (Padj) of 0.05 (n = 3 to 4 mice per genotype). (B) GO biological process analysis of Mll4-eKO differentially expressed down-regulated genes (1618 genes). Down-regulated genes driving the top GO term “lipoxygenase pathway” are highlighted. (C) Intersection of all MLL4-bound region-associated genes in NHEKs and down-regulated genes in Mll4-eKO mice (255 genes). (D) Left: GO biological process analysis of the intersection in (D) (255 genes). Right: List of key epidermal differentiation genes enriched in GO terms from (D) intersection. (E) ChIP-seq for MLL4 in proliferating or differentiated primary human keratinocytes at key epidermal differentiation (RARG) and barrier (SCNN1A) genes listed in Fig 2E (n = 2 to 3 per condition). (F) Immunoblot of Rarg and actin isolated from bulk epidermis of 3-week-old Mll4-eKO and control mice. Molecular ladder shown to the right of blot (n = 2 mice per genotype). (G) IHC staining of Sccn1a of 3-week-old Mll4-eKO and control mice epidermis (n = 3 mice per genotype. Scale bar, 200 μM). FDR, false discovery rate; FC, fold change.