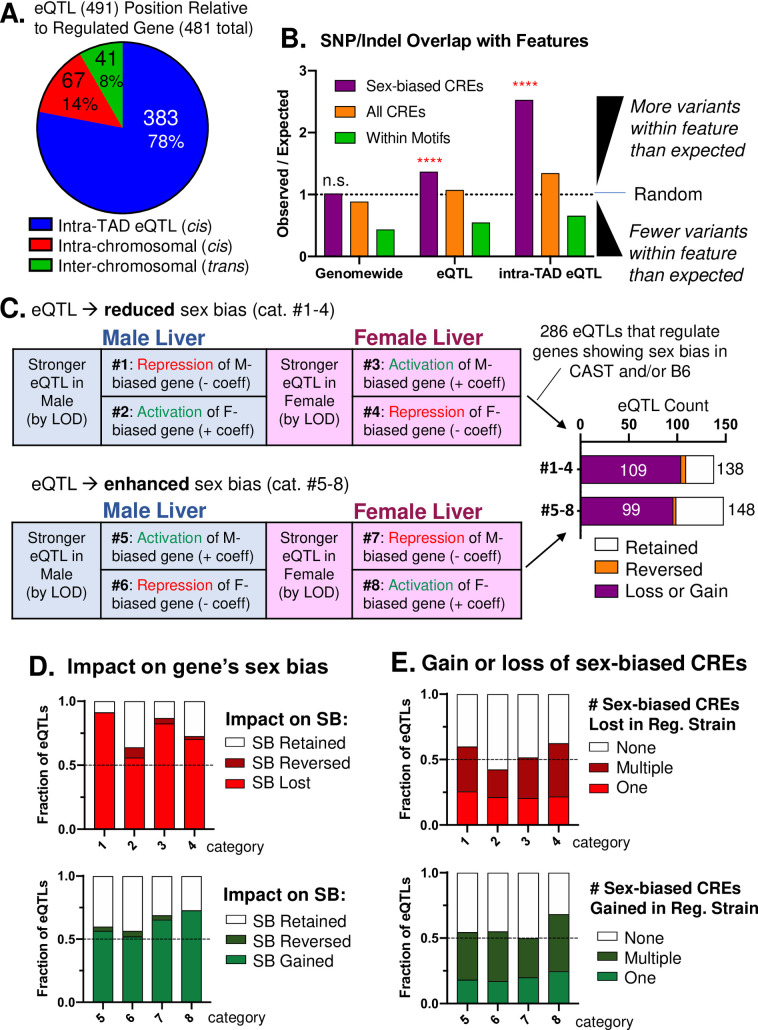
Fig 4. Genetic determinants of strain differences in sex-biased gene expression integrating DO-eQTLs and CAST/B6 founders.
A. eQTL position relative to its regulated gene target for the 491 eQTLs where CAST or B6 is the regulating strain. eQTLs were placed in three groups: regulated gene in the same TAD as the eQTL (n = 383); regulated gene on the same chromosome but not overlapping in the same TAD as the eQTL (n = 67); and regulated gene on a different chromosome than the eQTL (n = 41). B. Genetic variants are enriched for sex-biased CREs within eQTLs. Shown is the enrichment (observed/expected) of the number of genetic variants within any sex-biased CRE (purple), all CREs (orange), or within any TF-binding motif. The number of genetic variants overlapping the indicated feature (observed) was divided by the expected number of overlaps for a length-matched, peak genomic position-shuffled random control (median of 1,000 permutations). A value of 1 indicates no enrichment. Example: the expected number of strain-specific genetic variants in intra-TAD eQTLs (383 of 491 eQTLs) that overlap sex-biased CREs is 1,767 (number of strain-specific SNPs/Indels in the regulating strain overlapping by at least 1 bp randomized genomic positions matched in size and length to the set of sex-biased CREs; median across 1,000 permutations). In contrast, the observed number of strain-specific genetic variants in intra-TAD eQTLs that overlap the actual positions of sex-biased CREs is 4,463. Thus, Observed/Expected = 4,463/1,767 = 2.53. C. Categorization of the 388 sex-dependent eQTLs significant in only male or only female DO mice and that are stronger in either male or female liver (based on difference in LOD score). eQTL categories #1–4 result in the loss or reduction of sex bias, while eQTL categories #5–8 result in enhancement or gain of sex bias (figure based on [49]). 388 of the 491 eQTLs can be categorized into groups #1–8. Of these 388 eQTLs, 286 regulate genes that are sex-biased in either B6 or CAST RNA-seq (S9G and S9H Fig; complete listing in Sheet A in S6 Table). Right: number of eQTLs that result in a complete loss or de novo gain of sex-bias (purple; sex bias is significant in only CAST or B6), reversal of sex bias (orange; sex bias is significant in both CAST and B6, but in opposite directions), or where sex-bias is retained, but at either an increased or a reduced level (white bars). D. Impact on sex-biased gene expression for the 286 eQTLs in categories #1–8 that regulate genes with significant sex bias (SB) in B6 or CAST mouse liver. Plots show the fraction of eQTLs in each category that result in a gain/loss, reversal, or retain sex bias but at a reduced level, as in panel C. E. Fraction of the 286 eQTLs that contain one, multiple, or no sex-biased CREs with genetic variants that plausibly explain the gain/loss of sex-biased gene expression (relevant CREs, defined in Methods). Relevant CREs for categories #1–4 are those lost in the regulating strain (with the same directionality as the regulated gene); relevant CREs for categories #5–8 are those that are gained in the regulating strain (with the same directionality as the regulated gene). See listing in Sheet A in S6 Table. The fraction of eQTLs within each category that can be explained by relevant sex-biased and strain-specific CREs ranged from 42% (category #2) to 68% (category #8). Results are nearly identical when considering all 388 eQTLs in categories #1–8, 223 (57%) of which contain relevant gain or loss of sex-bias at CREs (Sheet A in S6 Table).