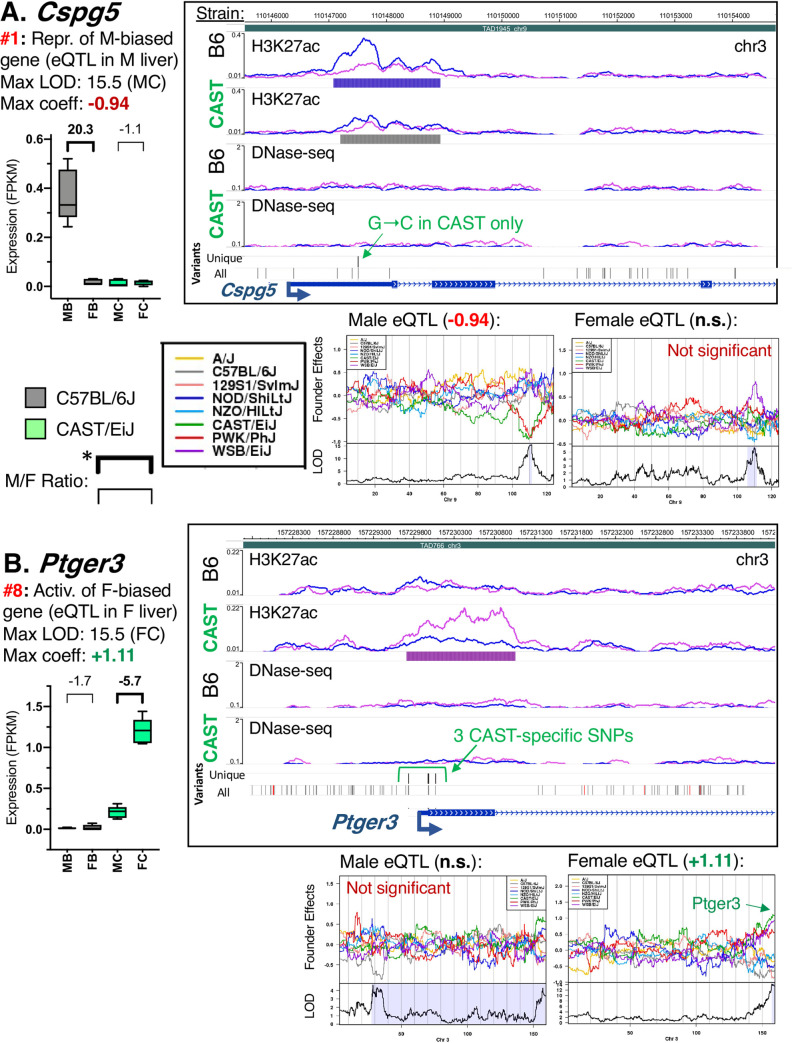
Fig 5. Single CREs within eQTL that result in loss of male bias or gain of female bias.
Data is shown for Cspg5 (A) and Ptger3 (B). The format used here is also used in Figs 6–8. Genes and eQTL categories (Fig 4C) are listed along with the strain, sex, and regression coefficient for the regulating strain. Box plots present expression data (in FPKM values, from polyA+ liver RNA-seq; Sheets G and H in S1 Table) across four mouse liver groups: male B6 (MB), female B6 (FB), male CAST (MC), and female CAST (FC) (n = 5 livers per group). Above each box is the linear fold-change, where positive values indicate male/female expression ratios and negative values indicate female/male expression ratios. Significant sex bias (FDR < 0.05) is indicated by bold. WashU Epigenome browser screenshot (right) includes these tracks (top to bottom): H3K27ac ChIP-seq signal with peak regions identified for B6 and CAST liver; DNase-seq signal and DHS, with peak regions identified for B6 and CAST liver; genetic variants that fall within CREs from the regulating strain (either CAST or B6, as indicated in summary; Unique: strain-specific variants; All, all variants between B6 and CAST liver), and RefSeq genes. For ChIP-seq and DNase-seq signal tracks, normalized male sequence reads are indicated by blue tracings, female reads by pink tracings (tracings superimposed within each strain). Peak and DHS tracks (horizontal bars below signal tracks) indicate male-bias (blue bars), female-bias (pink bars), and any robust peaks/DHS without significant sex bias (gray bars). See S5 Table for a complete listing of peaks and DHS. Below each screenshot: regression coefficient plots across the chromosome harboring the eQTL, shown for all 8 DO mouse founder strains, with the most relevant data shown in green (CAST) and gray (B6), as indicated in the box. Positive regression coefficient: genetic variants from that strain in the indicated region are associated with higher gene expression of the regulated gene; negative regression coefficient indicates the opposite effect. Regression coefficient values for the regulating strain are indicated above the plot, calculated separately for DO male (left) and DO female liver (right). n.s. indicates the association is not significant for any founder strain in the sex indicated. If CAST or B6 is not the regulating strain in a given sex this is indicated in red above the regression coefficient plot (Not significant in CAST or B6). A. The male-biased gene Cspg5 is repressed in CAST male liver, resulting in a loss of sex-biased expression in CAST liver (category #1 eQTL). This repression is associated with a single SNP (G to C in CAST) within the single male-biased peak in this eQTL that is lost in CAST mice (Sheet A in S6 Table). CAST is the regulating strain for this gene only in male DO mice (LOD = 15.5 in DO males; regression coefficient, -0.94). Significant male-biased expression is seen in B6 (20.3-fold M/F) but not in CAST liver. B. The female-biased gene Ptger3 is activated in CAST female liver, resulting in a gain of sex-biased expression in CAST liver (category #8 eQTL). This activation is associated with three SNPs, all within the single female-biased peak in this eQTL that is lost in B6 mice (Sheet A in S6 Table). CAST is the regulating strain for this gene only in female DO mice (LOD = 15.5 in DO males; regression coefficient, +1.11). Significant female-biased expression is seen in CAST (5.7-fold F/M) but not in B6 liver.