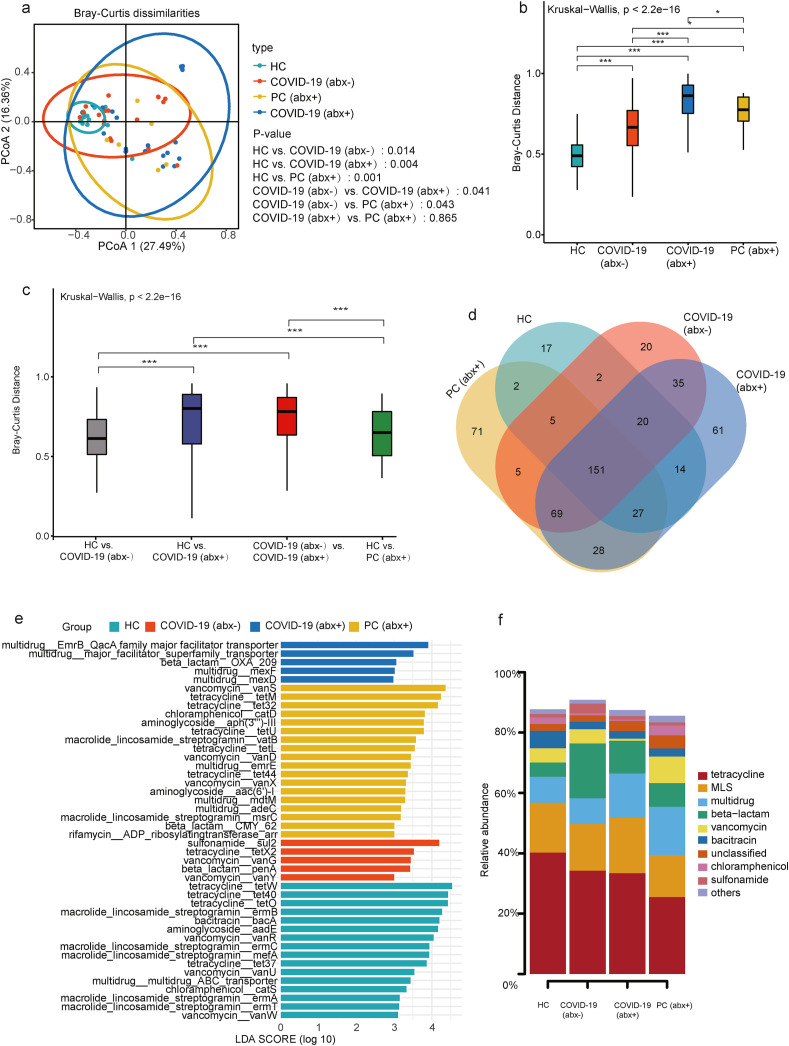
Fig. 2.
(a) Beta diversity was analyzed using principal coordinates analysis (PCoA) by Bray Curtis, showing segregation of the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+) group. Beta-diversity boxplots based on Bray-Curtis distances of metagenomics samples from the (b) same group (intra-beta diversity) and (c) between groups (inter-beta diversity). (d) Linear discriminant analysis effect size (LEfSe) histogram used to compare the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+) group. (e) Venn diagram showing shared and unique ARG subtypes amongst four groups. (f) Shared ARGs and abundance comparisons in the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+) group. In the stacked bar plot, the percentage (%) of a specific ARG in one of the four groups is equal to the ratio of its corresponding abundance to the sum of the abundances of the ARG in the four groups.