Extended Data Fig. 2. Source and coverage of available protein structures .
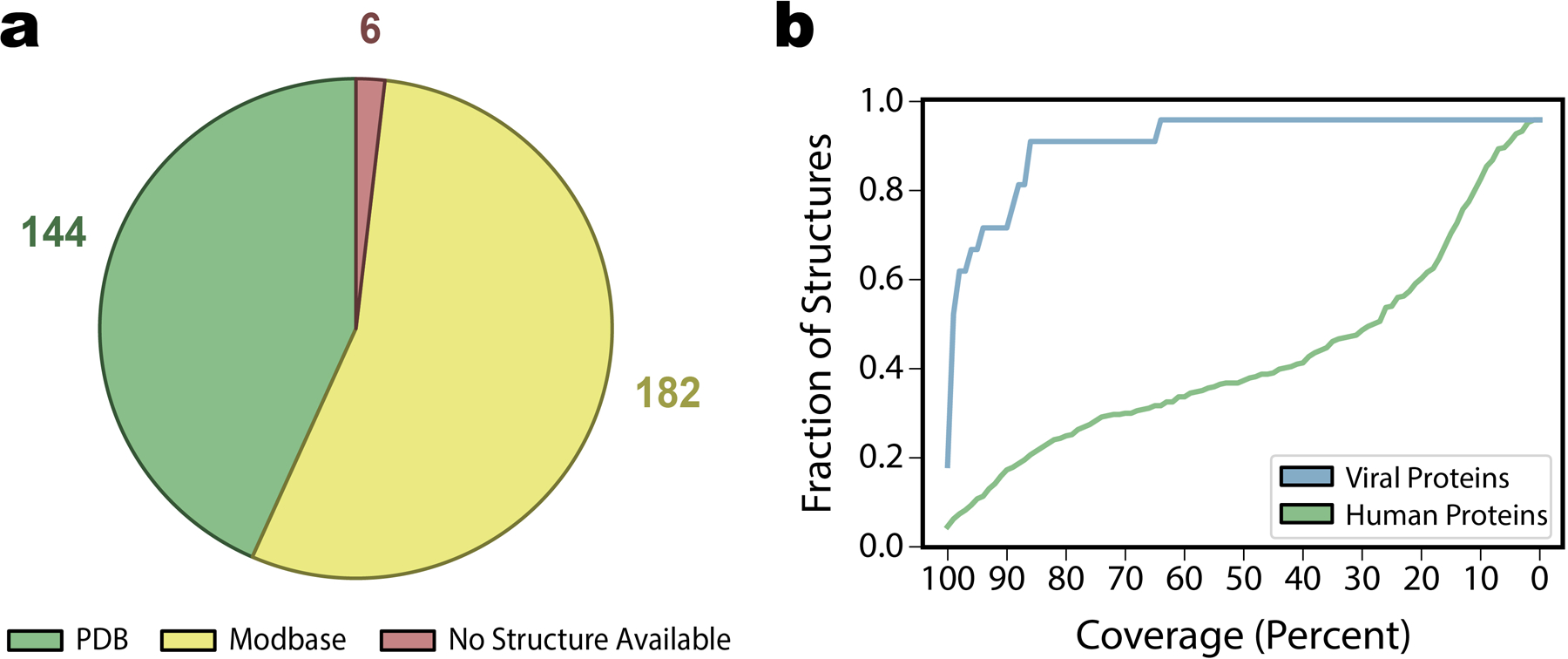
a, Breakdown of the source of all structures available for the 332 human interactors of SARS-CoV-2 as being either a experimentally solved structure from the Protein Data Bank (n=144) a homology model from Modbase (n=182), or no available structure (n=6). b, Analysis of the coverage of all available structures for both human (green) and viral (blue) proteins. The fraction of structures retained with coverage greater than or equal to a range of coverage thresholds is shown. For our purposes, all available structures were used for solvent accessibility feature calculations for ECLAIR predictions, but structures were only retained for docking if either 1) total coverage was at least 33% of 2) the structure covered at least one high-confidence interface prediction from ECLAIR.
