Figure 1. CHEK1 is identified as an ERKi sensitizer and essential for PDAC growth.
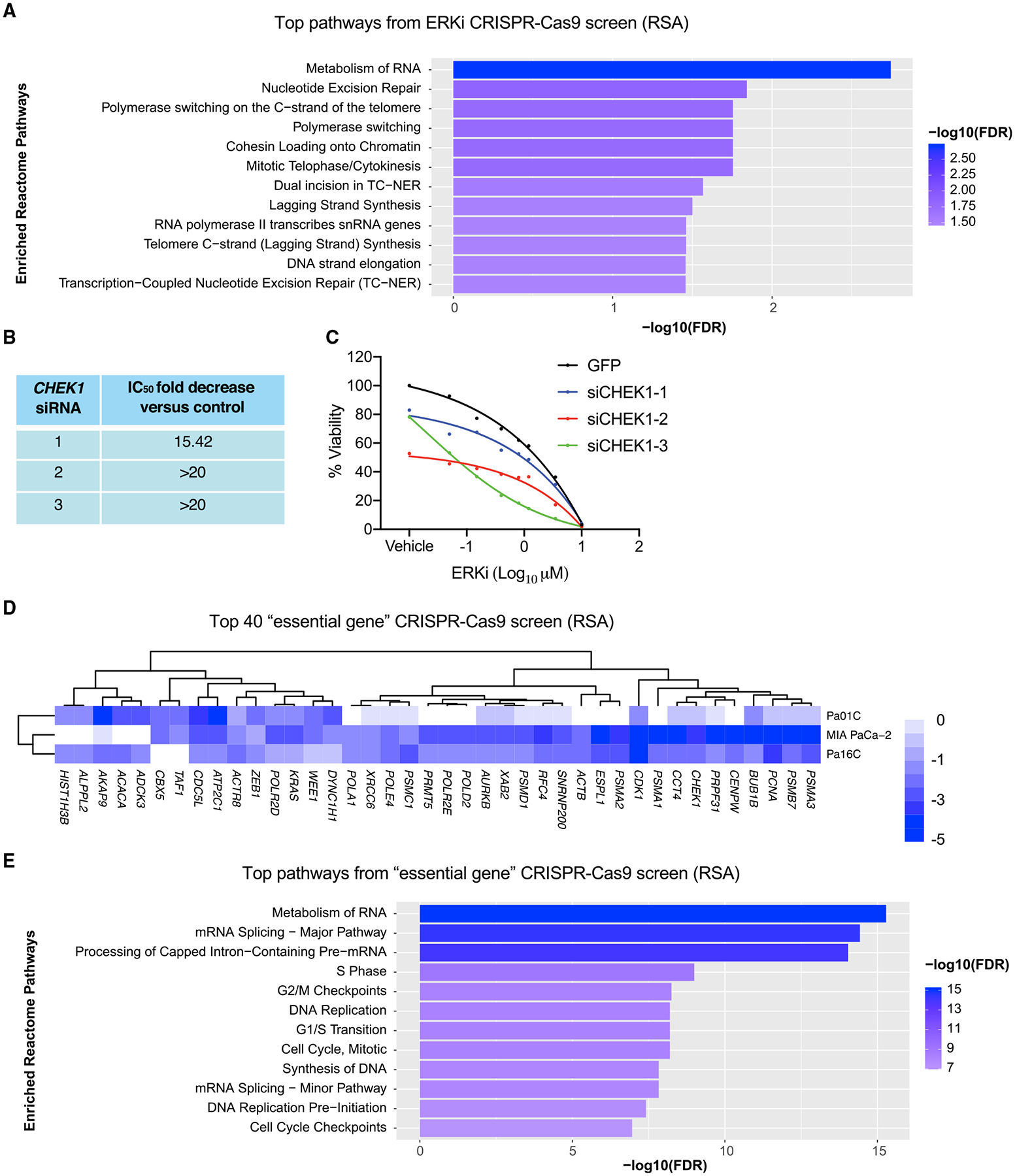
(A) Pathway analysis of the top 50 genes identified in a loss-of-function CRISPR-Cas9 screen (Pa01C, Pa14C, and PANC-1) targeting the druggable genome. Enriched ERKi sensitizer reactomes were determined using a STRING false discovery rate set at 5% and comparing the mean logP of all cell types and time points of the entire library.
(B) The shifts in ERKi GI50 with the top three CHEK1 siRNAs from the initial siRNA druggable genome screen.
(C) Viability curves (Pa16C) following 4 day treatment with ERKi and 5 day treatment with CHEK1 siRNA, with GFP siRNA as a control.
(D) The top 40 genes from the CRISPR-Cas9 “druggable genome” viability screen for identification of genes essential to PDAC cell growth; scale references the logP (RSA) value.
(E) Reactomes enriched with “essential” genes were identified using a STRING false discovery rate of 5% as described in (A).
