Fig. 3.
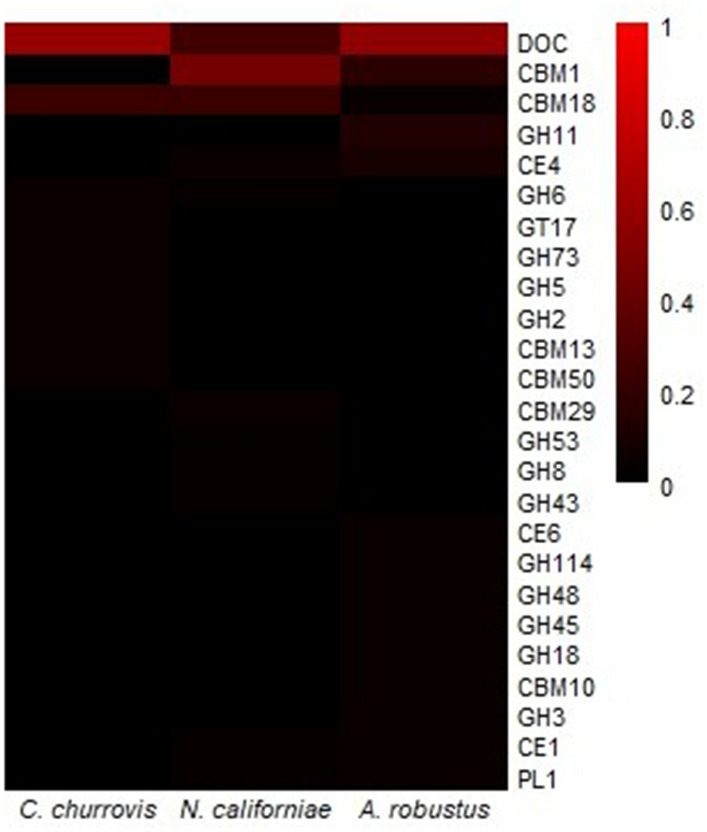
A heatmap of the proportion of genes containing domains belonging to CAZyme gene families or associated enzymatic machinery upregulated in co-cultures of three different fungal strains paired with the same non-native methanogen, Methanobacterium bryantii relative to fungal monocultures grown on a reed canary grass substrate. Three different strains of anaerobic fungi, Anaeromyces robustus, Neocallimastix californiae, and Caecomyces churrovis were used to form separate co-cultures with M. bryantii and grown on a reed canary grass substrate along with monocultures of each fungus on the same substrate. Differential expression of fungal genes in co-cultures relative to fungal monocultures was determined using DESEQ2. Gene domains were organized into the following classifications: carbohydrate binding modules (CBM), dockerins (DOC), glycoside hydrolases (GH), and glycosyltransferases (GT), polysaccharide lyases (PL), and carbohydrate esterases (CE)
