Abstract
Background
Obesity and related comorbidities are major health concerns among many US immigrant populations. Emerging evidence suggests a potential involvement of the gut microbiome. Here, we evaluated gut microbiome features and their associations with immigration, dietary intake, and obesity in 2640 individuals from a population-based study of US Hispanics/Latinos.
Results
The fecal shotgun metagenomics data indicate that greater US exposure is associated with reduced ɑ-diversity, reduced functions of fiber degradation, and alterations in individual taxa, potentially related to a westernized diet. However, a majority of gut bacterial genera show paradoxical associations, being reduced with US exposure and increased with fiber intake, but increased with obesity. The observed paradoxical associations are not explained by host characteristics or variation in bacterial species but might be related to potential microbial co-occurrence, as seen by positive correlations among Roseburia, Prevotella, Dorea, and Coprococcus. In the conditional analysis with mutual adjustment, including all genera associated with both obesity and US exposure in the same model, the positive associations of Roseburia and Prevotella with obesity did not persist, suggesting that their positive associations with obesity might be due to their co-occurrence and correlations with obesity-related taxa, such as Dorea and Coprococcus.
Conclusions
Among US Hispanics/Latinos, US exposure is associated with unfavorable gut microbiome profiles for obesity risk, potentially related to westernized diet during acculturation. Microbial co-occurrence could be an important factor to consider in future studies relating individual gut microbiome taxa to environmental factors and host health and disease.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-021-02559-w.
Keywords: Microbiome, Metagenomics, Hispanic population, Obesity
Background
Evidence from animal studies suggests that the gut microbiome may play a causal role in the development of obesity [1, 2], while the relationship between gut microbiome and obesity remains unclear in humans [3, 4]. Recent systematic reviews found mixed results on gut microbiome features associated with obesity across human studies [3, 4]. The inconsistency in human studies may be due to small sample sizes (the majority of previous studies had N < 100) and/or large interpersonal variation in gut microbiome related to many host factors (e.g., race/ethnicity [5], geography [6], diet/lifestyle [7, 8]). On the other side, microbes do not live in isolation but develop a range of relationships in the communities, including mutualism, commensalism, synergism, competition parasitism, predation, antagosim, and amensalism [9], and the complex microbial interaction might also influence the relationship of gut microbiome with obesity and other conditions.
Emerging evidence suggests that immigration is associated with gut microbiome alterations, which might be related to changes in diet and other factors during the acculturation process [10, 11]. Notably, microbiome-related conditions such as obesity and its comorbidities are major health concerns among US immigrants from lower-to-middle income countries [12]. A recent study in immigrants from Thailand to the USA found that US immigration was associated with loss of gut microbiome diversity and reduced ratio of Prevotella to Bacteroides, and loss of gut microbiome diversity was associated with obesity [10]. These results suggested a potential involvement of gut microbiota alterations in immigration-related obesity [10]. However, not much is known about individual taxonomic signals and functional components related to obesity. Higher Prevotella relative abundance has been consistently reported in non-industrialized populations, which are more metabolically healthy (e.g., lower prevalence of obesity and diabetes compared to industrialized populations), and whose diets contain more dietary fiber [13–15]. Paradoxically, several studies have found positive associations of Prevotella with obesity [16, 17], insulin resistance [18], and inflammatory autoimmune diseases [19, 20]. The mechanisms underlying the unexpected association between gut Prevotella and human diseases are not fully understood [21–23].
This report focuses on the US Hispanic/Latino community, which represent a substantial fraction of the total immigrant population in the USA (https://www.pewresearch.org/hispanic). In US Hispanics/Latinos, the prevalence of obesity is particularly high, and the length of duration of US residence or US exposure at earlier ages is associated with higher prevalence of obesity as well as greater severity of obesity [24, 25]. Our prior work using 16S rRNA data in 1674 US Hispanics/Latinos showed that the ratio of Prevotella to Bacteroides is lower in US-born versus immigrant Hispanics/Latinos, potentially related to lower fiber intake [11]. However, this ratio was found to be higher in individuals with obesity compared to those with normal weight, which was paradoxical to the observation that US-born Hispanics/Latinos had a higher prevalence of obesity and lower fiber intake compare to immigrant Hispanics/Latinos [11]. Therefore, studies are needed to explore potential explanations for these paradoxical results. Particularly, microbial interactions have been commonly observed [26, 27], but have not been well-considered in the analyses of gut microbiome and human health and disease. Microbial co-occurrence network which infer ecological relationships based on taxonomic composition data obtained from high-throughput sequencing techniques [28], has been used to visualize the correlations between microbes in microbial communities and predict the potential microbial interactions [9].
To better understand the interrelationship among immigration, gut microbiome, and obesity in US Hispanics/Latinos, we extended our analyses using shotgun metagenomics sequencing on a larger group of individuals. Among 2640 US Hispanics/Latinos of diverse background (including Dominican, Cuban, Puerto Rican, Mexican, Central American, and South American), aged 23 to 83 years, from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL ) [29, 30], we evaluated multiple gut microbiome features including community-level diversity, individual taxa, and functional potential, and examined associations of these features with immigration generation, duration of US residence, usual dietary intake, and obesity. The potential microbial co-occurrence relationship and its potential influences on the associations of gut microbiome with US immigration, dietary intake, and obesity were also examined.
Results
US exposure, obesity, and gut microbiome diversity
Among 2640 US Hispanics/Latinos (age ranged from 23 to 83 years), 368 (14%) were born in the mainland USA. Severity and prevalence of obesity were highest for those born in USA, as well as being positively associated with duration of US residence. Detailed study participant characteristics are shown in Additional file 1: Table S1.
Higher levels of obesity were associated with lower α-diversity Faith’s phylogenetic distance (PD) after multivariable adjustment in weighted linear regression (Additional file 1: Figure S1A). Consistently, α-diversity Faith’s PD index was lowest in those born in USA, and inversely associated with duration of US residence (Additional file 1: Figure S1A). Principal-coordinates analysis (PCoA) of weighted UniFrac distances indicated that both host obesity and US exposure (US born and duration of US residence) significantly co-varied with the β-diversity of gut microbiome and explained moderate proportion of variance (PERMANOVA analysis by adonis function from vegan package, R2 = 0.9% and 1.8% respectively, both P < 0.001) (Additional file 1: Figure S1B). These results are in line with previous findings in US immigrants from Thailand [10]. In addition, we also examined associations of other participant characteristics with β-diversity, and most variables were significantly associated with β-diversity, while all these R2 values were relatively small (R2 < 2.0%), including those for obesity and US exposure (Additional file 1: Figure S2). These results are consistent with those observed in our previous analyses using 16S data [11] as well as previous findings observed in large population-based studies in which multiple host factors showed significant associations with β-diversity but only explained a very small proportion of variation in β-diversity [6].
US exposure, obesity, and gut bacterial genera
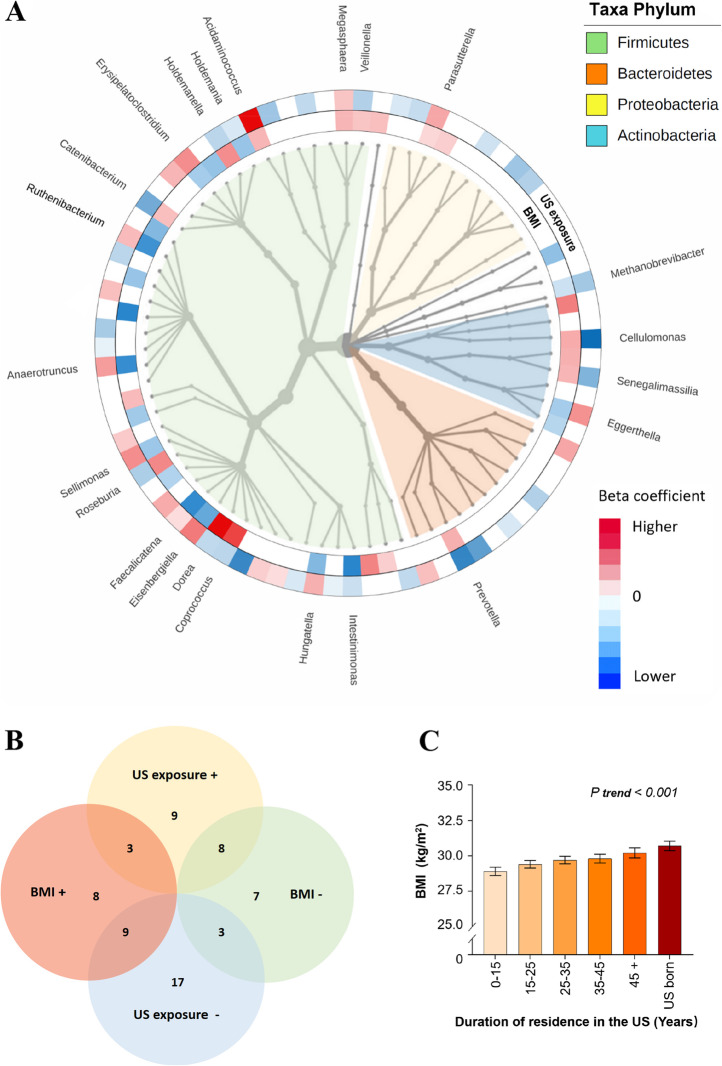
We then examined associations of individual gut microbial taxa at a genus level with obesity and US exposure after controlling for demographic, socioeconomic, behavioral, and clinical variables using weighted linear regression models. Of the 84 predominant gut bacterial genera included in this analysis (relative abundance ≥0.01%), 38 were significantly associated with BMI (FDR < 0.05; 26 under Firmicutes phylum, 6 under Actinobacteria, and 3 under Proteobacteria), and 49 genera were significantly associated with US exposure (FDR < 0.05; 33 under Firmicutes phylum, 6 under Proteobacteria, and 6 under Bacteroidetes) (Fig. 1A and Additional file 2: Table S2). Cross-classification of these two sets of results identified 23 bacterial genera that showed significant associations with both BMI and US exposure (Fig. 1A, B and Additional file 1: Table S3). Despite a strong positive association between BMI and US exposure (Fig. 1C), we found that a majority of genera (17 of 23) showed paradoxical associations with obesity and US exposure, including nine genera (e.g., Roseburia, Prevotella, and Dorea) associated with higher BMI but less US exposure, and the other eight genera (e.g., Anaerotruncus and Eggerthella) associated with lower BMI but greater US exposure. After further adjustment for Hispanic background, the associations of gut bacterial genera with obesity and US exposure did not change materially (Additional file 3: Table S4). In addition, in consideration of close correlations between US exposure and the covariates included in our regression models, we also compared associations of gut bacterial genera with US exposure and these covariates. Forty-nine genera were significantly associated with US exposure independently of these covariates, and their associations with these covariates are also shown in Additional file 4: Table S5.
Fig. 1.
Associations of gut bacterial taxa with host obesity and US exposure. A Integrated tree of microbial communities associated with host obesity and US exposure. Taxa from inner to outer circle represent bacteria kingdom to genus level; 84 predominant genera (average relative abundance > 0.01%) were included. The branch widths reflect the relative abundance of each detected taxon. Red/blue colors of the rings depict significant positive/inverse associations and the gradient colors reflect the ranks of effect sizes estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors, while white color indicates non-significant associations. The inner ring shows 38 genera significantly associated with host body mass index (BMI) (FDR P < 0.05), and the outer ring shows 49 genera significantly associated with US exposure (defined by place of birth and duration of residence in the US) (FDR P < 0.05). A total of 23 genera were significantly associated with both BMI and US exposure and the genera names are indicated. B Venn diagram of gut bacterial genera associated with BMI and US exposure. The symbol “+”/“−” depict significant positive/inverse associations in linear regression models (FDR P < 0.05). C Associations between host obesity and US exposure. Data are adjusted mean (SE) of BMI across US exposure groups estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors
To explore potential explanations for the observed paradoxical associations, we first examined these associations stratified by age, sex, geographic location, and Hispanic background as these factors may influence associations between gut microbiome features and human phenotypes. However, we did not find significant heterogeneity in associations of these bacterial genera with obesity (Additional file 1: Figure S3, and Additional file 5: Table S6) or US exposure (Additional file 3: Figure S4, and Additional file 6: Table S7) across strata. We also examined the associations of these bacterial genera with obesity and US exposure among the first-generation Hispanic/Latino immigrants according to the place of birth (i.e., Dominica, Cuba, Puerto Rica, Mexico, Central America, and South America), and did not find significant heterogeneity in these associations (Additional file 7: Table S8 and Additional file 8: Table S9). In addition, we performed sensitivity analyses by excluding individuals with antibiotic use during the past 6 months, and the results on the associations of gut bacteria genera with obesity and US exposure were similar to those of the primary analyses after adjustment for antibiotic use in the regression models. These results suggest that these host factors are less likely to explain the observed paradoxical associations.
We also examined a total of 88 species within these 23 genera but found little evidence suggesting that variation in species levels explain the observed paradoxical associations.
Results at the genus level were mostly driven by one or two major species under corresponding genera (Additional file 1: Table S10). For example, Prevotella copri, the most predominant species in Prevotella genus (mean relative abundance: 11%, accounting for 88.3% of Prevotella species), was associated with higher BMI but less US exposure.
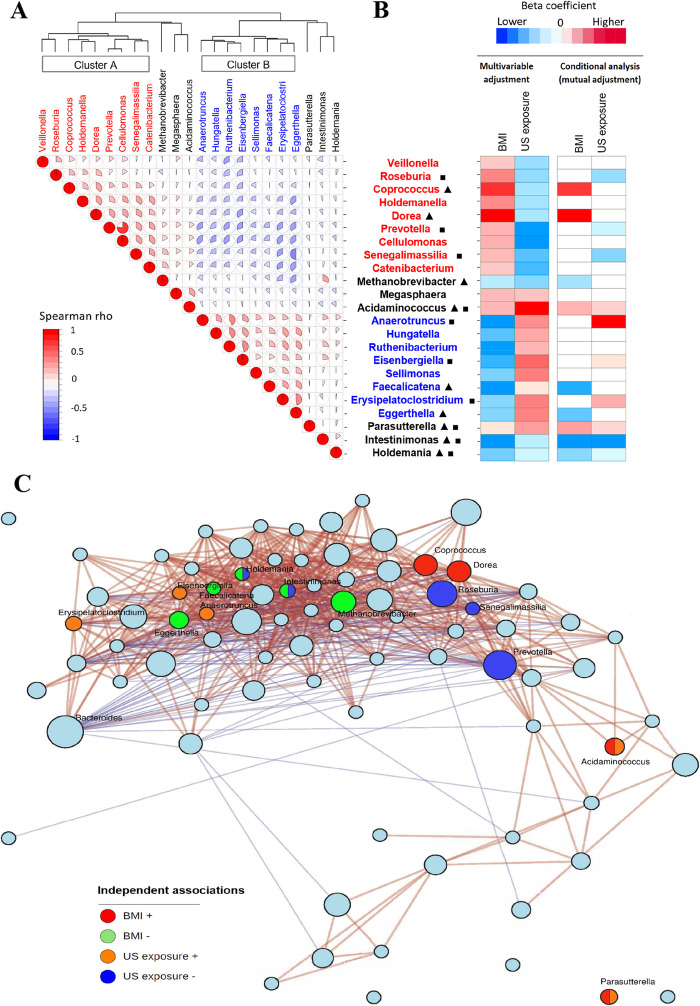
We then examined correlations among these 23 genera associated with both obesity and US exposure. Given the compositional data, we applied multiple methods including Spearman, Pearson and SparCC to calculate correlation coefficients among bacteria genera [31], which showed generally consistent results (Additional file 1: Figure S5). As gut microbiome data tend to be zero-inflated, this may influence the correlation estimates. Thus, we performed sensitivity analyses after excluding samples with absent bacteria genera, and similar correlations among these genera were observed (Additional file 1: Figure S5). The 17 bacterial genera which showed paradoxical associations formed two clusters which were inversely correlated (Fig. 2A). Cluster A included correlated bacteria (Spearman r = 0.06–0.77) that were associated with higher BMI and less US exposure. Cluster B included correlated bacteria (Spearman r = 0.18–0.46) that were associated with lower BMI and greater US exposure.
Fig. 2.
Relationships among the identified gut microbial genera associated with both obesity and US exposure. A Spearman correlation heatmap for the 23 identified bacterial genera associated with both obesity and US exposure. B Associations of 23 gut bacteria genera with obesity and US exposure. These associations were estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors (multivariable adjustment, FDR P < 0.05 level) and further adjustment for other bacterial genera (conditional analysis, mutual adjustment). Red/blue colors depict significant positive/inverse associations and the gradient colors reflect the ranks of effect sizes estimated in linear regression models, while white color indicates non-significant associations. Triangle and square indicate the microbial genera that were independently associated with obesity and US exposure after mutual adjustment, respectively. C Co-occurrence network representing the structure of microbial composition. The size of node reflects the cumulative sum scaling normalized abundance of each taxon. Red/green nodes indicate genera positively/inversely associated with host obesity after mutual adjustment (P < 0.05), and orange/ blue nodes indicate genera positively/inversely associated with US exposure after mutual adjustment (P < 0.05). Positive (red lines) and inverse (blue lines) correlations were obtained between predominant genera using Spearman rho based co-occurrence network analysis
Given the moderate correlations among many of these genera, we hypothesized that the observed paradoxical associations might be due to correlations/co-occurrence among these genera. To test this hypothesis, we performed conditional analyses including all genera associated with both BMI and US exposure in the same model, except for Cellulomonas which was excluded due to its high correlation with Prevotella (r = 0.77), to identify potentially independent bacterial taxa associated with obesity and/or US exposure. After controlling for other genera, nine genera remained significantly associated with obesity, and 10 genera remained significantly associated with US exposure (Fig. 2B, and Additional file 1: Table S11). No paradoxical associations remained after conditional analyses. Among six bacteria outside of clusters A and B, Acidaminococcus and Parasutterella were associated both with higher BMI and greater US exposure, while Intestinimonas and Holdemania were associated with lower BMI and less US exposure. Among nine members of Cluster A, Coprococcus and Dorea were associated with higher BMI only, and Roseburia, Prevotella and Senegalimassilia were associated with less US exposure only. Among eight members of Cluster B, Faecalicatena and Eggerthella were associated with lower BMI only, and Anaerotruncus, Eisenbergiella, and Erysipilatoclostria were associated with greater US exposure only. To minimize potential influences of over-adjustment, we conducted conditional analyses which only included four independent BMI-associated genera (Coprococcus, Dorea, Faecalicatena, and Eggerthella) in the models. The associations of other 13 genera in clusters A or B with BMI were not significant after adjustment for these four independent BMI-associated genera (Additional file 1: Table S11). Similarly, the associations of other 11 genera in cluster A or B with US exposure were not significant after adjustment for six independent US exposure-associated genera. In addition, we examined the associations of these genera with two other obesity measurements, body fat percentage, and waist circumference (Additional file 1: Figure S6), as well as severity of obesity (normal weight, 18.5 ≤ BMI < 25 kg/m2; overweight, 25 ≤ BMI < 30 kg/m2); obese I, 30 ≤ BMI < 35 kg/m2; obese II, 35 ≤ BMI < 40 kg/m2; and obese III, BMI ≥ 40 kg/m2). The results on the associations of these genera with body fat percentage, waist circumference, and severity of obesity in the conditional analyses were generally consistent with those on the associations between these genera and BMI (Additional file 1: Figure S6, and Additional file 1: Table S12).
In order to explore the relationship among microbial genera in a compositionally coherent manner, we also applied differential ranking analysis using songbird pipeline [32] in which the natural log-ratio of microbial genera data was calculated using Clostridium as the reference. We built a Spearman correlation heatmap using these log-ratio data (Additional file 1: Figure S6), and the correlations among microbial genera were generally consistent with those estimated using the CSS transformed data. For examples, we also observed significant positive correlations among Coprococcus, Roseburia, Prevotella, and Dorea, suggesting potential bacterial co-occurrence of these genera.
We also performed co-occurrence network analysis [33, 34] to visualize potential bacterial co-occurrence relationships among gut bacterial genera in our study samples. This analysis indicated potential bacterial co-occurrence relationships among these genera which showed associations with both obesity and US exposure (Fig. 2C), and their associations might be driven by some key genera associated with obesity or US exposure. For example, among Cluster-A bacterial genera which showed bacterial co-occurrence relationships, Coprococcus and Dorea were associated with higher BMI, while Roseburia, Prevotella, and Senegalimassilia were associated with less US exposure. Interestingly, either phylogenetically closely related bacterial genera, or those belonging to different phyla, could be clustered in the bacterial co-occurrence network. For example, Roseburia, Coprococcus, and Dorea are in the Lachnospiraceae family, while Prevotella and Senegalimassilia belong to the Bacteroidetes phylum and the Actinobacteria phylum, respectively (Fig. 1A). Taken together, these results suggested that the observed associations of individual gut microbial taxa with obesity and US exposure might be due to bacterial co-occurrence, and these paradoxical associations were resolved after taking bacterial co-occurrence into account.
US exposure, dietary intake, and gut microbiome
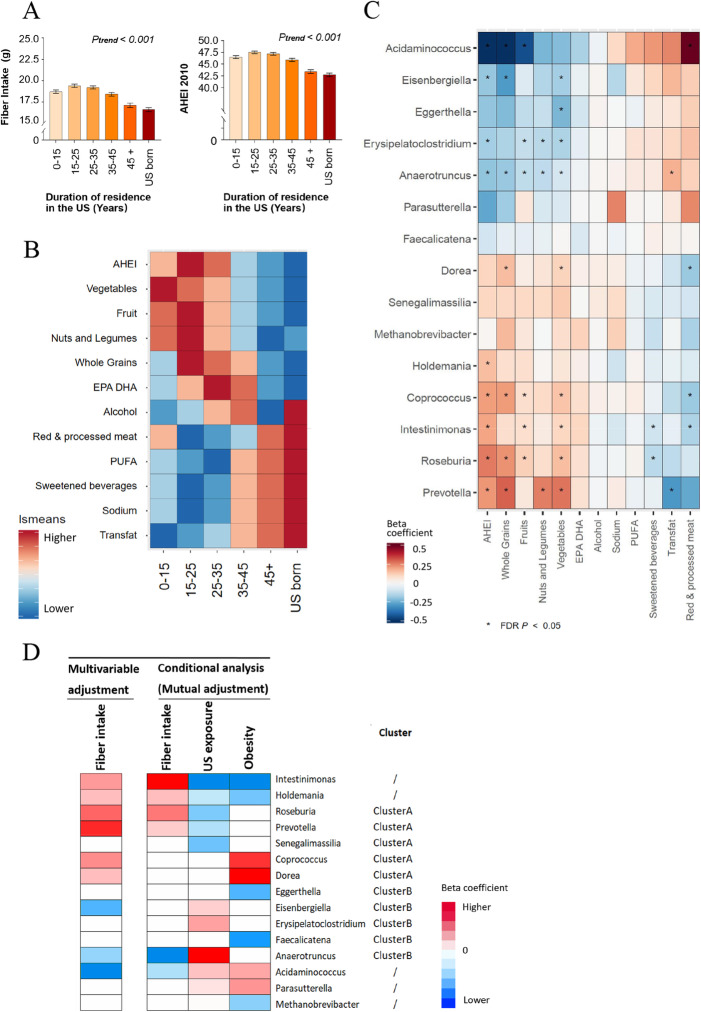
To further clarify the relationships of individual gut microbial taxa with obesity and US immigration, we attempted to account for diet, a behavioral factor that tends to change after immigration and that can profoundly influence both body habitus and gut microbiome features [11]. Compared to US-born Hispanics/Latinos, Latin America-born individuals, especially those who had relative shorter duration of US residence, showed higher consumption of dietary fiber, and generally more favorable eating habits, as evidenced by higher levels of the 2010-AHEI as well as individual food/nutrient components (e.g., higher intakes of vegetables, fruits, nuts, and legumes, and lower intakes of red/processed meat, sugar-sweetened beverages, sodium, and trans-fat) (Fig. 3A, B).
Fig. 3.
US exposure, dietary intake, and gut microbial genera. A Associations of US exposure with dietary fiber intake and AHEI-2010 score. Data are adjusted means (SEs) of dietary fiber intake and AHEI-2010 score across US exposure groups estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors. B Heatmap for overall dietary quality and individual dietary factor associated with US exposure. Gradient colors reflect the ranks of adjusted means of AHEI-2010 score and 11 food/nutrition component of AHEI-2010 across US exposure groups estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors. C Heatmap for overall dietary quality and individual dietary factor associated with gut microbial genera. This heatmap includes 15 microbial genera that were independently associated with obesity and/or US exposure. Data are effect sizes of dietary factors on gut microbial genera estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors. D Associations of gut microbiota with dietary fiber intake, US exposure, and host obesity. These associations were estimated after adjustment for demographic, socioeconomic, behavioral, and clinical factors (multivariable adjustment, FDR P < 0.05 level) and further adjustment for other bacterial genera (conditional analysis, mutual adjustment). Red/blue colors depict significant positive/inverse associations and the gradient colors reflect the ranks of effect sizes estimated in linear regression models, while white color indicates non-significant associations
We then related individual bacteria genera with host dietary intake and found that 30 out of 84 predominant bacteria genera were significantly associated with 2010 -AHEI as well as individual food/nutrient components, especially with healthy fiber-rich foods (e.g., whole grain, vegetables, fruits, nuts, and legumes) (Additional file 1: Figure S8). Consistent with the relationship between US exposure and host dietary intake, bacterial genera associated with less US exposure were associated with relatively healthier diet pattern and higher intakes of fiber-rich foods, while bacterial genera associated with greater US exposure were associated with relatively unhealthier diet pattern and lower intakes of fiber-rich foods, regardless of their associations with obesity (Fig. 3C). We then focused on the associations of these bacteria genera with dietary fiber intake and found some unexpected associations. Particularly, Dorea and Coprococcus, which were independently associated with obesity, were associated with higher fiber intake (Fig. 3D). After including all 15 bacteria genera in a conditional analysis model (mutual adjustment) on fiber intake, no paradoxical associations remained. Six bacteria genera were independently associated with dietary fiber intake, and directions of these associations were in line with those of their associations with US exposure and/or obesity (Fig. 3D).
US exposure, fiber intake, and gut microbiome functional components
Lastly, we examined microbial function contents to better understand the interrelationship among US immigration, dietary intake, gut microbiome, and obesity. The gut microbiome functional profiles were obtained using SHOGUN [35] and annotated by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database Release 94.0 [36], and a total of 1952 known enzymes with specific Enzyme Commission numbers (EC number) were identified and included in the analyses. We first examined associations between US exposure and KO groups for 1952 known enzymes using weighted linear regression models and identified 260 KO groups associated with US exposure (all FDR P < 0.1). We then performed an enrichment test for these KO groups at EC level II enzyme category and found an enrichment of enzymes belonging to the glycosylases associated with US exposure (Additional file 1: Table S13).
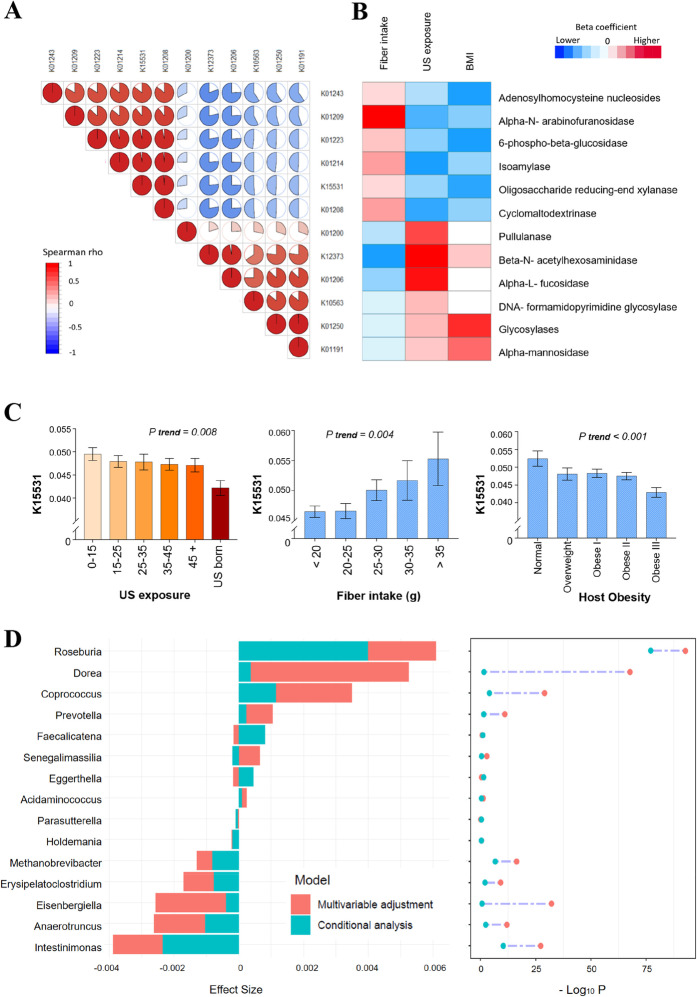
We thus focused on the glycosylases and found 12 KO groups associated with US exposure (all FDR P < 0.1, Fig. 4A; and Additional file 1: Table S14). These KO groups formed two clusters and KO groups within each cluster were highly correlated with each other (Fig. 4A). The first cluster which was associated with less US exposure included KO groups encoding enzymes related to degradation of dietary fibers. For example, oligosaccharide reducing-end xylanase (K15531; EC3.2.1.156) was known as a high molecular mass xylanases which can degrade xylan, a type of dietary fiber found in plant cell walls [37, 38]. Consistently, these KO groups were associated with higher fiber intake. The second cluster which was associated with greater US exposure included KO groups encoding enzymes related to starch degradation (e.g., Pullulanase, K01200, EC3.2.1.41) or carbohydrate metabolism associated with obesity and insulin resistance (e.g., alpha- mannosidase, K01191, EC3.2.1.24) [39]. These results suggest that, compared to those with a shorter duration of US residence, US-born Hispanics/Latinos and those with a longer duration of US residence may have gut microbiota with decreased capacity for fiber degradation and increased capacity for starch degradation and simple carbohydrate utilization, potentially related to decreased consumption of fiber-rich foods and increased consumption of refined grains and sugary foods. We then related these KO groups with BMI and found expected associations showing that all KO groups associated with less US exposure and higher fiber intake were associated with lower BMI, and some KO groups associated with greater exposure and lower fiber intake were associated with higher BMI (Fig. 4B). For example, levels of oligosaccharide reducing-end xylanase (K15531) deceased with greater US exposure, increased with higher fiber intake, and deceased with higher BMI/severity of obesity (Fig. 4C). The inverse association between oligosaccharide reducing-end xylanase and US exposure might be mainly driven by relatively lower levels of oligosaccharide reducing-end xylanase in US-born Hispanics/Latinos compared to immigrant Hispanics/Latinos, as we only found a trend for the inverse association between oligosaccharide reducing-end xylanase and years of US residence among immigrant Hispanics/Latinos (P = 0.10).
Fig. 4.
Gut microbial functional components associated with both US exposure and dietary fiber intake, and their associations with obesity and gut bacterial genera. A Correlation heatmap for 12 microbial functional enzymes associated with both US exposure and dietary fiber. B Association of 12 microbial functional enzymes with US exposure, dietary fiber intake and BMI. These associations were estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors. Red/blue colors depict significant positive/inverse associations (FDR P < 0.05) and the gradient colors reflect the ranks of effect sizes estimated in linear regression models, while white color indicates non-significant associations. C Levels of gut microbial enzyme K15531 xylanase according to US exposure, dietary fiber intake, and host obesity. Data are adjusted mean (SE) of enzyme K15531 xylanase (centered log-ratio transformed) across US exposure groups, levels of dietary fiber intake, and host obesity status estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors. D Associations of 15 gut bacterial genera with gut microbial enzyme K15531 xylanase. Data are effect sizes and P values estimated in linear regression models after adjustment for demographic, socioeconomic, behavioral, and clinical factors (multivariable adjustment) and further adjustment for other bacterial genera (conditional analysis, mutual adjustment)
We then explored potential contributions of these 15 bacterial genera that were independently associated with US exposure, fiber intake, and/or obesity to these KO groups. As these KO groups were highly correlated with each other, we focused on xylanase (K15531) as an example, given its well-known biological function in fiber degradation. While we found expected positive associations between bacterial genera (e.g., Roseburia and Prevotella which were associated with higher fiber intake) and xylanase, Dorea and Coprococcus which were positively associated with obesity were also positively associated with xylanase (Fig. 4D). We performed conditional analysis by including all 15 genera in the same regression model on xylanase. This greatly attenuated or abolished most of the associations of these genera, especially Dorea and Coprococcus, with xylanase, while the association between Roseburia and xylanase was only slightly attenuated and remained the strongest (Fig. 4D). These results suggest that Roseburia might be a major bacteria contributing to xylanase, and the observed positive associations of Dorea and Coprococcus with xylanase might be due to their correlations with Roseburia. Our genomic analyses provided further evidence supporting the presence of xylanase gene on representative Roseburia genomes but not on representative Dorea or Coprococcus genomes (Additional file 1: Table S15). In support of our results, Roseburia species from the human gut have been reported to produce xylanases and display high xylanolytic activity [38].
Discussion
Taken together, these data from a large population-based cohort study of Hispanics/Latinos living in the US demonstrated that US immigration was associated with alterations in gut microbiome diversity, individual taxa, and functional profiles, which is consistent with previous findings observed in immigrants from Thailand to the USA [10]. Further extending prior work, this study linked these US immigration-associated gut microbiome features with obesity, a major health burden among US Hispanics/Latinos and other US immigrants from lower-to-middle income countries [12]. In particular, we focused on individual gut microbial taxa associated with both US immigration and obesity. We identified several gut bacterial genera associated with US immigration, host dietary intake, and obesity in expected directions, suggesting a potential involvement of these bacteria linking US immigration with obesity, though potential mechanisms underlying these associations remain unknown. For example, Acidaminococcus which was associated with obesity in the current study and other populations [40, 41] was found to be more abundant in US-born Hispanics/Latinos and those with a longer duration of US residence, compared to those with a shorter duration of US residence, potentially related to increased consumption of animal-based protein-rich foods (e.g., red/processed meat intake; Fig. 3B), since Acidaminococcus is known to use amino acids as energy source [42]. However, it remains unknown how increased gut Acidaminococcus may influence host obesity.
Another major contribution of this study is to demonstrate that considering potential microbial co-occurrence may help clarify the paradoxical associations of individual gut microbial taxa with US immigration and obesity. Our current analysis suggested that the observed associations of some gut microbioal taxa with US immigration or obesity might be due to their co-occurrence with other taxa. For example, we found that Roseburia along with Prevotella were the most common genera associated with US immigration in this population, and their paradoxical associations with obesity might be due to their correlations with Dorea and other key obesity-associated taxa. Unlike Prevotella, the influence of immigration on Roseburia was not reported before, although Roseburia is known to degrade and use xylans and other dietary fibers [38, 43, 44] and the potential beneficial role of Roseburia in health has also been suggested [43, 45]. Our data indicate that Roseburia, like Prevotella, might be another gut microbiome signature of non-industrialized populations and might decrease during westernization. On the other hand, the paradoxical association between Dorea, an obesity-associated bacterial genus in ours and other studies [46, 47], and less US exposure might be related to Roseburia and other key taxa associated with US immigration. Although it is unknown how Dorea may influence host obesity, our data suggested that this might not be related to US immigration.
It has been suggested that high strain-level diversity of Prevotella might explain the inconsistent associations of Prevotella with human health and disease across populations [22, 23]. However, there are scant data on Prevotella strains in relation to human disease traits given the difficulties of strain-level analysis in large human population-based studies. A recent study reported that Prevotella copri, the most abundant species of Prevotella in the human gut, may encompass four distinct clades, but there was little evidence suggesting these four clades to be differentially associated with obesity, diabetes, or other human diseases [21]. Our study provides another potential explanation. The inconsistent associations between Prevotella and human diseases (e.g., obesity) might be due to variation in correlations between Prevotella and some true disease-associated taxa across populations. For example, the observed association between Prevotella and obesity in this US Hispanic/Latino population might be due to the correlation between Prevotella and Dorea, an obesity-associated bacterial genus. However, more studies are needed to clarify whether the inconsistent associations between Prevotella and human diseases are due to strain-level diversity of Prevotella, correlations between Prevotella and disease-associated bacteria, or both.
The observed bacterial co-occurrence relationships might reflect some positive bacterial interactions that generate increased benefits for the group, such as mutualism (e.g., antibiotic resistance conferral), commensalism (e.g., cross-feeding on compounds produced by other members), and synergism (e.g., syntrophic cooperation) [9]. Our results are also supported by the previously reported co-occurrence relationship between Coprococcus and Roseburia, both in the Lachnospiraceae family [34, 48]. These data suggest that the phylogenetically closely related genera could also have co-occurrence relationship rather than always compete against each other due to their similar habitat preference [34]. However, due to the poor mechanistic understanding of the human gut microbial community, the ecological links among gut bacteria should be interpreted with caution. Although the underlying mechanisms for the bacterial co-occurrence network need to be elucidated, our findings may have important implications in the future studies investigating associations of gut microbiome with human health and disease. We have shown that the associations of individual taxa with environmental factors (e.g., US exposure) and host phenotypes (e.g., obesity) could be confounded by other taxa due to potential bacterial co-occurrence, and thus, conditional analysis using mutual adjustment model is needed to examine the independent associations of individual taxa with environmental factors and host phenotypes. Furthermore, microbial co-occurrence network analysis [28] might be a useful approach to examine the bacterial co-occurrence relationship and its potential influences on the associations between gut microbiome and human health and disease.
This study has several limitations. Data on diet were assessed prior to the gut microbiome assessment, although we obtained dietary information using rigorous methods based on two 24-h dietary recalls and a food propensity questionnaire designed to capture habitual long-term diet [49] and detected strong associations between host dietary intake and gut microbiome features. The potential urban-rural differences could not be evaluated in our study due to the lack of detailed information on place of origin. Due to limitations of shallow shotgun metagenomics, we were unable to examine strain-level data and determine individual taxa contributing to gut microbiome functional components using SHOGUN pipeline [35]. Given the observational nature of this study, causal inference could not be established without further evidence.
Conclusions
In conclusion, this study in US Hispanics/Latinos found that US immigration was associated with reduced gut microbiome diversity, reduced gut microbiome functions of fiber degradation, and alterations in individual gut microbial taxa (e.g., increased Acidaminococcus, decreased Roseburia, and Prevotella), potentially related to westernized diet during acculturation process. The US immigration-related alterations in gut microbiome features were also associated with obesity, suggesting a potential role of gut microbiome in the development of obesity among US immigrants from Latin America. Of note, a majority of gut bacterial genera showed paradoxical associations with US immigration, host dietary intake, and obesity, which could be resolved into directionally consistent associations after accounting for potential microbial co-occurrence. Thus, potential microbial co-occurrence may be an important factor to consider in future studies relating individual gut microbial taxa to environmental factors and host health and disease.
Methods
Study population
HCHS/SOL is a prospective, population-based cohort study of 16,415 Hispanic/Latino adults (aged 18–74 years at the time of recruitment during 2008–2011) who were selected using a two-stage probability sampling design from randomly sampled census block areas within four US communities (Chicago, IL; Miami, FL; Bronx, NY; San Diego, CA) [11, 29, 30]. A comprehensive battery of interviews and a clinical assessment with blood draw were conducted at in-person clinic visits. Information on demographics, behaviors, health status, family and medical histories, and medication use was collected using structured questionnaires, and blood pressure and anthropometric traits were measured. Blood biomarkers including blood lipids and glycemic traits were measured by standard methods [50]. The HCHS/SOL Gut Origins of Latino Diabetes (GOLD) ancillary study was conducted to examine the role of gut microbiome in the development of multiple health outcomes, enrolling 3035 participants from the HCHS/SOL approximately concurrent with the second in-person visit period from 2014 to 2017 [11]. This study included participants of diverse Hispanic/Latino backgrounds, including Dominican, Cuban, Puerto Rican, Mexican, Central American, and South American. The study was approved by the institutional review boards of corresponding site institutions. Written informed consent was obtained from all participants.
Fecal specimen collection and Shotgun metagenomics sequencing
Enrolled GOLD participants were provided with a stool collection kit. For each participant, a single fecal specimen was self-collected using a disposable paper inverted hat (Protocult collection device, ABC Medical Enterprises, Inc., Rochester, MN). Two fecal samples were self-collected by each participant, with one sample collected by a Whatman FTA card (GE Healthcare, Chicago, IL) and another one placed in a tube of RNAlater (Invitrogen, Carlsbad, CA). Detailed procedures have been described elsewhere [11]. In addition, use of antibiotics or probiotic supplements within the prior 6 months and stool characteristics were ascertained by self-administered questionnaire at the time of stool sample collection.
Metagenomics Sequencing was performed on DNA extracted from fecal samples collected by FTA card using a novel shallow-coverage method of shotgun sequencing-based Illumina NovaSeq platforms [51]. The adapters and barcode indices are processed following the iTru adapter protocol [52]. De-multiplexing was applied to generate Shallow shotgun per-sample FASTQ data and the adapter sequences were trimmed. The human-filtered FASTQ reads were further trimmed to remove low-quality bases that had a PHRED quality score of 25 or less using prinseq-lite 0.20.4 (https://edwards.sdsu.edu/cgi-bin/prinseq/prinseq.cgi) [53]. For the final analytical set, two Illumina sequencing runs were pooled. Samples with a coverage depth less than 500,000 reads per sample were excluded. Of 3035 samples, 2640 samples passed all QC metrics and were used in the current analysis. The coverage depth ranged from 500 to 8945 k reads per sample and average depth was 955 k reads per sample. The quality-controlled paired end data was then concatenated and aligned against the NCBI RefSeq representative prokaryotic genome collection (release 82) [54] using default SHOGUN [35] settings. Bowtie2 [55] was selected as the alignment tools in SHOGUN pipeline (https://github.com/knights-lab/SHOGUN). The reads that mapped to a single reference genome is labeled with the NCBI taxonomic annotation at the species level. Those reads that mapped to multiple reference genomes are labeled as the last common ancestor (LCA) of each label according to the NCBI taxonomy [35]. The α-diversity indices (Faith’s phylogenetic distance), and β-diversity weighted UniFrac distances were calculated using Qiita (https://qiita.ucsd.edu/) [56], Metaphlan3(https://huttenhower.sph.harvard.edu/metaphlan/) [57], and R phyloseq / vegan packages (https://github.com/joey711/phyloseq/https://github.com/vegandevs/vegan) [58, 59]. Functional profiles were obtained using SHOGUN and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database release 94.0 [36].
Assessment of US exposure, obesity, and dietary intake
Information on Hispanic/Latino background, place of birth, and years living in the mainland USA (with the US territory of Puerto Rico considered to be part of Latin America) was collected using structured questionnaires [11, 29, 30]. US exposure was defined based on place of birth and duration of US residence: mainland US born, US residence of 0–< 15 years, US residence of 15–< 25 years, US residence of 25–< 35 years, US residence of 35–< 45 years, and US residence of ≥ 45 years. Body mass index (BMI) was calculated as measured weight (kg) divided by measured height squared (m2). Obesity status was defined by BMI: normal weight (18.5 ≤ BMI < 25 kg/m2), overweight (25 ≤ BMI < 30 kg/m2), obese I (30 ≤ BMI < 35 kg/m2), obese II (35 ≤ BMI < 40 kg/m2), and obese III (BMI ≥ 40 kg/m2). Other obesity measures included waist circumference, and body fat percentage obtained from the Tanita body composition analyzer (model TBF-300A; Tanita Corporation, Arlington Heights, IL).
Usual dietary intake was estimated using the National Cancer Institute methodology based on dietary data collected from two 24-h dietary recalls and a food propensity questionnaire (FPQ), as described previously [60]. In brief, the first dietary recall was administered through in-person interviews conducted at the time of the baseline visit, whereas the second was performed primarily via telephone approximately 30 days after the first interview. The FPQ, which was administered at the 1-year follow-up call, asked participants to report frequencies of foods eaten in the previous year. Foods and nutrients were analyzed using the multiple-pass methods of the Nutrition Data System for Research software (version 11) from the Nutrition Coordinating Center at University of Minnesota. Overall dietary quality for each participant was estimated by the 2010 Alternative Healthy Eating Index (AHEI-2010) which was established based on extensive epidemiologic findings linking foods and nutrients to chronic disease outcomes [61]. The AHEI-2010 consists of 10 food/nutrient components in addition to alcohol consumption, with each of the 11 components being given 0–10 points according to predetermined criteria.
Statistical analysis
Clinical and demographic characteristics of the study population were described by reporting means with standard deviations (SDs) or medians with interquartile ranges for continuous variables and absolute frequencies with percentages for categorical variables.
Weighted linear regression models were applied to examine associations of microbial α-diversity indices (Faith’s phylogenetic distance) with BMI and US exposure, adjusting for age, sex, study center, education level, income, antibiotic use in the last 6 months, moderate-to-vigorous physical activity, total energy intake, diabetes, and metformin use. Permutational multivariate analysis of variance (PERMANOVA) and principal-coordinate analysis (PCoA) were carried out with weighted UniFrac distances for the microbial β-diversity analyses.
For metagenomics taxonomic analyses, a total of 84 predominant bacterial genera with average relative abundance ≥0.01% were included (a total of 596 species belonged to these predominant genera). Cumulative sum scaling (CSS) normalization was conducted [62] for each of taxonomic variables before analyses. Associations of bacterial taxonomic features with BMI and US exposure were examined using weighted multivariable linear regression, with adjustment for the aforementioned covariates. Furthermore, we further adjusted for Hispanic background in additional multivariable regression models. All the models incorporated the sampling weights from the complex survey design of the HCHS/SOL [29, 30]. We constructed integrated hierarchical tree using iTol (https://itol.embl.de/) [63].
In order to explore the relationship among microbial genera in a compositionally coherent manner, we applied differential ranking (DR) analysis using songbird pipeline (https://github.com/biocore/songbird ) [32]. Read counts were summarized at the genus level and inputted into Songbird [32] for multinomial regression. To ensure proper model fit while guarding against potential model overfitting, a null model was generated using similar parameters. Comparing this null model to the fitted model demonstrated better fit for the latter (pseudo-Q2 = 0.022301), enabling further utilization of the differentials [32]. These differentials were then inputted into Qurro for visualization [64]. Then we selected the genus Clostridium as the reference, since it was stable (with minimum absolute ranks) across experimental conditions (BMI) and present across most samples. The natural log-ratio of microbial genera data was calculated using Clostridium as the reference and we also built Spearman correlation heatmap using these log-ratio data.
We examined associations of bacterial genera with BMI and US exposure stratified by sex, age group dichotomized at the median age of study participants (age < 55 or ≥ 55 years), study center (Chicago, Miami, Bronx, or San Diego), Hispanic/Latino background (6 strata: Dominican, Cuban, Puerto Rican, Mexican, Central American, and South American; 2 strata: Mainland or Caribbean), place of birth (US born or non-US born; BMI analysis only), and obesity status (obese or non-obese; US exposure analysis only). We also examined the associations of bacterial genera with obesity and US exposure among the first-generation Hispanic/Latino immigrants according to the place of birth (i.e., Dominica, Cuba, Puerto Rica, Mexico, Central America, and South America). In addition, we performed sensitivity analyses by excluding individuals with antibiotic use during the past 6 months.
To identify potential independent bacterial genera associated with BMI or US exposure, we included all 23 genera which were associated with both BMI and US exposure in the same regression model on BMI or US exposure (mutual adjustment, conditional analysis). We applied multiple methods including Spearman, Pearson, and SparCC to calculate correlation coefficients among bacteria genera [31]. In addition, we performed hierarchical clustering approach to identify the clusters. Microbial co-occurrence network, which infer ecological associations based on taxonomic composition data obtained from high-throughput sequencing techniques [28], are widely used to visualize the statistically significant associations between microbes in microbial communities and predict the potential microbe interactions [9]. In this study, we performed Spearman rho based co-occurrence network analysis with Cytoscape (https://cytoscape.org/) [33], to explore the relationships among the identified bacterial taxa and to better describe the overall structure of microbial components. Nodes and edges represent bacterial genera and statistically significant correlations between these genera, respectively.
Associations of dietary fiber intake, AHEI-2010 score, and 11 food and nutrient components of AHEI-2010 with US exposure and 84 predominant gut bacterial genera were examined using weighted multivariable linear regressions, with adjustment for the aforementioned covariates. The conditional analysis with mutual adjustment further included 15 identified genera which were independently associated with BMI and/or US exposure, to further examine independent associations between bacterial taxa and dietary fiber intake.
For metagenomics functional analyses, centered log-ratio transformation was applied to the KEGG ortholog group abundances. Weighted linear regression models were applied to examine associations of KEGG ortholog groups with US exposure, dietary fiber intake, and BMI, after controlling for the aforementioned covariates, and incorporating the sampling weights from the complex survey design. An enrichment analysis using Fisher’s exact test was performed for the 1952 annotated enzymes at EC level II enzyme category. Partial spearman correlation analysis was use to estimate correlation coefficients between bacterial genera (CSS normalized) and KO groups (centered log-ratio transformed). Associations of 15 genera which were independently associated with obesity and/or US exposure with xylanase (K15531) were examined using linear regression models after multivariable adjustment. We further included all 15 genera in the same model (mutual adjustment, conditional analysis) to examine potential independent associations of these genera with xylanase (K15531).
The Benjamini-Hochberg false discovery rate (FDR) method was used for multiple testing correction. Statistical analyses were performed using R 3.6.1. unless otherwise stated.
Supplementary Information
Additional file 1: Figure S1. Microbial community diversity associated with obesity and US exposure. Figure S2. Beta diversity PERMANOVA analysis for obesity, US exposure, and other co-variates, ranked by R2 Figure S3. Associations of gut microbiota and host obesity (BMI), stratified by: A. Sex; B. Age group (< 55 and > = 55 years); C. Place of birth (US born and non-US born); D. Hispanic origin (Caribbean [Dominican, Cuban, Puerto Rican] and Mainland [Central American, Mexican, South American]) ; E to J. Field Center (Bronx, Chicago, Miami, San Diego). Figure S4. Associations of gut microbiota and US exposure, further stratified by : A. Sex; B. Hispanic origin (Caribbean [Dominican, Cuban, Puerto Rican] and Mainland [Central American, Mexican, South American]); C. Obesity; D. Age group (< 55 and > = 55 years); E to J. Field Center (Bronx, Chicago, Miami, San Diego). Figure S5. Correlation heatmap for the 23 identified bacterial genera associated with both obesity and US exposure. Figure S6. Associations of 23 gut microbial genera with BMI, Body fat percentage and waist circumference. Figure S7. Correlation heatmap for all bacterial genera, using log ratios data and Clostridium as reference. Figure S8. Heatmap for overall dietary quality and individual dietary factor associated with gut microbial genera. Table S1. Characteristics of the study population by US exposure Table S3. Multivariable regression analyses: 23 genera associated with both obesity and US exposure. Table S10. Association of host obesity and US exposure with gut microbiota: species level analyses. Table S11. Conditional analysis for genera associated with both obesity and US exposure Table S12. Levels of gut microbial genera according to host obesity. Table S13. Target US exposure associated enzyme selection: Enrichment test for the annotated enzymes Table S14. Regression analyses: Enrichment test selected key enzymes associated with both US exposure and fiber intake. Table S15. The presence of key enzymes in specific bacteria: Alignment analysis.
Additional file 2: Table S2. Associations of 84 gut bacterial genera with obesity and US exposure.
Additional file 3: Table S4. Associations of 84 gut bacterial genera with obesity and US exposure after further adjusted for Hispanic background.
Additional file 4: Table S5. Associations of 84 gut bacterial genera with US exposure and all covariates in the multivariable regression model.
Additional file 5: Table S6. Associations of gut bacterial genera with obesity (BMI), stratified by Hispanic background groups.
Additional file 6: Table S7. Associations of gut bacterial genera with US exposure, stratified by Hispanic background groups.
Additional file 7: Table S8. Associations of gut bacterial genera with obesity (BMI), stratified by birth place, among the first generation immigrants.
Additional file 8: Table S9. Associations of gut bacterial genera with US exposure, stratified by birth place, among the first generation immigrants.
Peer review information
Kevin Pang was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 9.
Authors’ contributions
Study concept and design: ZW, QQ, and RCK. Acquisition of data: YVB, DM, ZW, MU, RK, RDB, RCK, and QQ. Analysis and interpretation of data: ZW, RCK, and QQ. Drafting of the manuscript: ZW, RCK, and QQ. Critical revision of the manuscript for important intellectual content: All authors. Obtained funding: RCK, RDB, QQ, KN and RK. Study supervision: RCK and QQ. The authors read and approved the final version of the manuscript.
Funding
The Hispanic Community Health Study/Study of Latinos (HCHS/SOL) is a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (HHSN268201300001I/N01-HC-65233), University of Miami (HHSN268201300004I/N01-HC-65234), Albert Einstein College of Medicine (HHSN268201300002I/N01-HC-65235), University of Illinois at Chicago – HHSN268201300003I/N01-HC-65236 Northwestern University), and San Diego State University (HHSN268201300005I/N01-HC-65237). The following Institutes/Centers/Offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. Additional funding for the “Gut Origins of Latino Diabetes” (GOLD) ancillary study to HCHS/SOL was provided by 1R01MD011389-01 from the National Institute on Minority Health and Health Disparities. Other funding sources for this study include R01-DK119268, R01-DK120870, the New York Regional Center for Diabetes Translation Research (P30DK111022) from the NIDDK, and R01-HL060712, R01-HL140976, and R01-HL136266 from the NHLBI. None of the funding agencies had a role in the design, conduct, interpretation, or reporting of this study.
Availability of data and materials
HCHS/SOL data are archived at the National Institutes of Health repositories dbGap (accession number phs000810.v1.p1) and BIOLINCC (accession number HLB01141418a), and gut microbiome sequence data in this study are deposited in QIITA, ID 11666, and EMBL-EBI ENA, ERP117287 [65]. HCHS/SOL has established a process for the scientific community to apply for access to participant data and materials, with such requests reviewed by the project’s Steering Committee. These policies are described at https://sites.cscc.unc.edu/hchs/ (accessioned November 19, 2021). The corresponding author will accept reasonable requests for data and specimen access, which will be referred to the Steering Committee of the HCHS/SOL project.
Declarations
Ethics approval and consent to participate
All participants enrolled in the HCHS/SOL completed informed consent at the time of enrollment into the cohort and subsequently provided informed consent to participate in the gut microbiome ancillary study project. IRBs of all participating institutions approved the study. The IRB of the lead institution (Albert Einstein College of Medicine) has approved the HCHS/SOL project under reference number 2007-432. The National Institutes of Health maintains an Observational Study Monitoring Board which reviews the project, participant safety, and burden. All experimental methods comply with the Helsinki Declaration.
Consent for publication
All participants provided written informed consent for collection, analysis, and publication of their study data and results of laboratory tests derived from their biospecimens. The manuscript was reviewed by the HCHS/SOL Publications Committee which provided its approval for submission of this work for publication
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zheng Wang, Email: zheng.wang@einsteinmed.org.
Mykhaylo Usyk, Email: mykhaylo.usyk@einsteinmed.org.
Yoshiki Vázquez-Baeza, Email: yoshiki89@gmail.com.
Guo-Chong Chen, Email: gcchen@suda.edu.cn.
Carmen R. Isasi, Email: carmen.isasi@einsteinmed.org
Jessica S. Williams-Nguyen, Email: jswn@uw.edu
Simin Hua, Email: siminhua03@gmail.com.
Daniel McDonald, Email: d3mcdonald@ucsd.edu.
Bharat Thyagarajan, Email: thya0003@umn.edu.
Martha L. Daviglus, Email: daviglus@uic.edu
Jianwen Cai, Email: cai@bios.unc.edu.
Kari E. North, Email: kari_north@unc.edu
Tao Wang, Email: tao.wang@einsteinmed.org.
Rob Knight, Email: robknight@ucsd.edu.
Robert D. Burk, Email: robert.burk@einsteinmed.org
Robert C. Kaplan, Email: robert.kaplan@einsteinmed.org
Qibin Qi, Email: qibin.qi@einsteinmed.org.
References
- 1.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 2.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science. 2010;328(5975):228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sze MA, Schloss PD. Looking for a signal in the noise: revisiting obesity and the microbiome. MBio. 2016;7. 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed]
- 4.Crovesy L, Masterson D, Rosado EL. Profile of the gut microbiota of adults with obesity: a systematic review. Eur J Clin Nutr. 2020;74(9):1251–1262. doi: 10.1038/s41430-020-0607-6. [DOI] [PubMed] [Google Scholar]
- 5.Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, Tremaroli V, Bakker GJ, Attaye I, Pinto-Sietsma SJ, van Raalte DH, Snijder MB, Nicolaou M, Peters R, Zwinderman AH, Bäckhed F, Nieuwdorp M. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med. 2018;24(10):1526–1531. doi: 10.1038/s41591-018-0160-1. [DOI] [PubMed] [Google Scholar]
- 6.He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, Chen MX, Chen ZH, Ji GY, Zheng ZDX, Mujagond P, Chen XJ, Rong ZH, Chen P, Lyu LY, Wang X, Wu CB, Yu N, Xu YJ, Yin J, Raes J, Knight R, Ma WJ, Zhou HW. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018;24(10):1532–1535. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 7.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 9.Ma B, Wang Y, Ye S, Liu S, Stirling E, Gilbert JA, Faust K, Knight R, Jansson JK, Cardona C, Röttjers L, Xu J. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome. 2020;8(1):82. doi: 10.1186/s40168-020-00857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, et al. US immigration westernizes the human gut microbiome. Cell. 2018;175:962–972.e10. doi: 10.1016/j.cell.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan RC, Wang Z, Usyk M, Sotres-Alvarez D, Daviglus ML, Schneiderman N, et al. Gut microbiome composition in the Hispanic community health study/study of Latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol. 2019;20:219. doi: 10.1186/s13059-019-1831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Commodore-Mensah Y, Selvin E, Aboagye J, Turkson-Ocran RA, Li X, Himmelfarb CD, et al. Hypertension, overweight/obesity, and diabetes among immigrants in the united states: an analysis of the 2010-2016 national health interview survey. BMC Public Health. 2018;18:773. doi: 10.1186/s12889-018-5683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, Fiori J, Gotti R, de Bellis G, Luiselli D, Brigidi P, Mabulla A, Marlowe F, Henry AG, Crittenden AN. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5(1):3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thingholm LB, Rühlemann MC, Koch M, Fuqua B, Laucke G, Boehm R, et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe. 2019;26:252–264.e10. doi: 10.1016/j.chom.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu HJ. Park SG, Jang HB, Choi MK, Park KH, Kang JH, et al. Obesity alters the microbial community profile in Korean adolescents. PLoS One. 2015;10(7):e0134333. doi: 10.1371/journal.pone.0134333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 19.Wen C, Zheng Z, Shao T, Liu L, Xie Z, Le Chatelier E, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 2017;18:142. doi: 10.1186/s13059-017-1271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. 2019;26:666–679.e7. doi: 10.1016/j.chom.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol. 2016;13(2):69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 23.De Filippis F, Pasolli E, Tett A, Tarallo S, Naccarati A, De Angelis M, et al. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe. 2019;25:444–453.e3. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Creighton MJ, Goldman N, Pebley AR, Chung CY. Durational and generational differences in Mexican immigrant obesity: is acculturation the explanation? Soc Sci Med. 2012;75(2):300–310. doi: 10.1016/j.socscimed.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isasi CR, Ayala GX, Sotres-Alvarez D, Madanat H, Penedo F, Loria CM, Elder JP, Daviglus ML, Barnhart J, Siega-Riz AM, van Horn L, Schneiderman N. Is acculturation related to obesity in Hispanic/Latino adults? Results from the Hispanic community health study/study of Latinos. J Obes. 2015;2015:186276. doi: 10.1155/2015/186276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, van Kleunen M, Becks L, Thakur MP. Towards a general understanding of bacterial interactions. Trends Microbiol. 2020:783–5. [DOI] [PubMed]
- 27.Scheuerl T, Hopkins M, Nowell RW, Rivett DW, Barraclough TG, Bell T. Bacterial adaptation is constrained in complex communities. Nat Commun. 2020;11:754. doi: 10.1038/s41467-020-14570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirano H, Takemoto K. Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinformatics. 2019;20(1):329. doi: 10.1186/s12859-019-2915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavange LM, Kalsbeek WD, Sorlie PD, Aviles-Santa LM, Kaplan RC, Barnhart J, et al. Sample design and cohort selection in the Hispanic community health study/study of latinos. Ann Epidemiol. 2010;20(8):642–649. doi: 10.1016/j.annepidem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorlie PD, Aviles-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, et al. Design and implementation of the Hispanic community health study/study of Latinos. Ann Epidemiol. 2010;20(8):629–641. doi: 10.1016/j.annepidem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y, et al. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 2016;10(7):1669–1681. doi: 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, et al. Establishing microbial composition measurement standards with reference frames. Nat Commun. 2019;10:2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otasek D, Morris JH, Bouças J, Pico AR, Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20:185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Guo Z, Xue Z, Sun Z, Zhang M, Wang L, Wang G, Wang F, Xu J, Cao H, Xu H, Lv Q, Zhong Z, Chen Y, Qimuge S, Menghe B, Zheng Y, Zhao L, Chen W, Zhang H. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME Journal. 2015;9(9):1979–1990. doi: 10.1038/ismej.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, et al. Evaluating the information content of shallow shotgun metagenomics. mSystems. 2018;3. 10.1128/mSystems.00069-18 eCollection 2018 Nov-Dec. [DOI] [PMC free article] [PubMed]
- 36.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T, Yang Y, Liang Y, Jiao X, Zhao C. Beneficial effect of intestinal fermentation of natural polysaccharides. Nutrients. 2018. 10.3390/nu10081055. [DOI] [PMC free article] [PubMed]
- 38.Mirande C, Kadlecikova E, Matulova M, Capek P, Bernalier-Donadille A, Forano E, Béra-Maillet C. Dietary fibre degradation and fermentation by two xylanolytic bacteria bacteroides xylanisolvens XB1A and Roseburia intestinalis XB6B4 from the human intestine. J Appl Microbiol. 2010;109(2):451–460. doi: 10.1111/j.1365-2672.2010.04671.x. [DOI] [PubMed] [Google Scholar]
- 39.Lee S, Zhang C, Kilicarslan M, Piening BD, Bjornson E, Hallström BM, Groen AK, Ferrannini E, Laakso M, Snyder M, Blüher M, Uhlen M, Nielsen J, Smith U, Serlie MJ, Boren J, Mardinoglu A. Integrated network analysis reveals an association between plasma mannose levels and insulin resistance. Cell Metab. 2016;24(1):172–184. doi: 10.1016/j.cmet.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugino KY, Paneth N, Comstock SS. Michigan cohorts to determine associations of maternal pre-pregnancy body mass index with pregnancy and infant gastrointestinal microbial communities: late pregnancy and early infancy. PLoS One. 2019;14(3):e0213733. doi: 10.1371/journal.pone.0213733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborne G, Wu F, Yang L, Kelly D, Hu J, Li H, Jasmine F, Kibriya MG, Parvez F, Shaheen I, Sarwar G, Ahmed A, Eunus M, Islam T, Pei Z, Ahsan H, Chen Y. The association between gut microbiome and anthropometric measurements in Bangladesh. Gut Microbes. 2020;11(1):63–76. doi: 10.1080/19490976.2019.1614394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vital M, Penton CR, Wang Q, Young VB, Antonopoulos DA, Sogin ML, et al. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome. 2013;1:8. doi: 10.1186/2049-2618-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.La Rosa SL, Leth ML, Michalak L, Hansen ME, Pudlo NA, Glowacki R, et al. The human gut firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat Commun. 2019;10:905. doi: 10.1038/s41467-019-08812-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chassard C, Bernalier-Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol Lett. 2006;254(1):116–122. doi: 10.1111/j.1574-6968.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- 45.Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, Bonnaure-Mallet M, Jolivet-Gougeon A. Roseburia spp.: a marker of health? Future Microbiol. 2017;12(2):157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 46.Ottosson F, Brunkwall L, Ericson U, Nilsson PM, Almgren P, Fernandez C, Melander O, Orho-Melander M. Connection between BMI-related plasma metabolite profile and gut microbiota. J Clin Endocrinol Metab. 2018;103(4):1491–1501. doi: 10.1210/jc.2017-02114. [DOI] [PubMed] [Google Scholar]
- 47.Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X, Xia H, Liu Z, Cui B, Liang P, Xi L, Jin J, Ying X, Wang X, Zhao X, Li W, Jia H, Lan Z, Li F, Wang R, Sun Y, Yang M, Shen Y, Jie Z, Li J, Chen X, Zhong H, Xie H, Zhang Y, Gu W, Deng X, Shen B, Xu X, Yang H, Xu G, Bi Y, Lai S, Wang J, Qi L, Madsen L, Wang J, Ning G, Kristiansen K, Wang W. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23(7):859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Zheng J, Shi W, Du N, Xu X, Zhang Y, et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut. 2018;67(9):1614–1625. doi: 10.1136/gutjnl-2018-315988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattei J, Sotres-Alvarez D, Daviglus ML, Gallo LC, Gellman M, Hu FB, Tucker KL, Willett WC, Siega-Riz AM, van Horn L, Kaplan RC. Diet quality and its association with cardiometabolic risk factors vary by Hispanic and Latino ethnic background in the Hispanic community health study/study of latinos. J Nutr. 2016;146(10):2035–2044. doi: 10.3945/jn.116.231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi Q, Strizich G, Merchant G, Sotres-Alvarez D, Buelna C, Castaneda SF, et al. Objectively measured sedentary time and cardiometabolic biomarkers in US Hispanic/Latino adults: the Hispanic community health study/study of latinos (HCHS/SOL) Circulation. 2015;132(16):1560–1569. doi: 10.1161/CIRCULATIONAHA.115.016938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costello M, Fleharty M, Abreu J, Farjoun Y, Ferriera S, Holmes L, et al. Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genomics. 2018;19:332. doi: 10.1186/s12864-018-4703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glenn TC, Nilsen RA, Kieran TJ, Sanders JG, Bayona-Vásquez NJ, Finger JW, Pierson TW, Bentley KE, Hoffberg SL, Louha S, Garcia-de Leon FJ, del Rio Portilla MA, Reed KD, Anderson JL, Meece JK, Aggrey SE, Rekaya R, Alabady M, Belanger M, Winker K, Faircloth BC. Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext) PeerJ. 2019;7:e7755. doi: 10.7717/peerj.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vazquez-Baeza Y, Ackermann G, et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods. 2018;15(10):796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with biobakery 3. eLife. 2021;10:e65088. doi: 10.7554/eLife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMurdie PJ, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oksanen J. Multivariate analysis of ecological communities in R: Vegan tutorial. version 2.3-0. 2015; https://cran.r-project.org/web/packages/vegan/vegan.pdf.
- 60.Siega-Riz AM, Sotres-Alvarez D, Ayala GX, Ginsberg M, Himes JH, Liu K, Loria CM, Mossavar-Rahmani Y, Rock CL, Rodriguez B, Gellman MD, van Horn L. Food-group and nutrient-density intakes by Hispanic and Latino backgrounds in the hispanic community health study/study of Latinos. Am J Clin Nutr. 2014;99(6):1487–1498. doi: 10.3945/ajcn.113.082685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiuve SE, Fung TT, Rimm EB, Hu FB, McCullough ML, Wang M, et al. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr. 2012;142(6):1009–1018. doi: 10.3945/jn.111.157222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10(12):1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letunic I, Bork P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fedarko MW, Martino C, Morton JT, González A, Rahman G, Marotz CA, et al. Visualizing 'omic feature rankings and log-ratios using qurro. NAR Genom Bioinform. 2020;2:lqaa023. doi: 10.1093/nargab/lqaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaplan RC,Wang Z, Usyk M, Sotres-Alvarez D, Daviglus ML, Schneiderman N, et al. Burk_SOL GOLD. ERP117287. EMBL-EBI European Nucleotide Archive. https://www.ebi.ac.uk/ena/data/search?query=ERP117287.ID11666.QIITA . https://qiita.ucsd.edu/study/description/11666#. Accessed 19 Nov 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Microbial community diversity associated with obesity and US exposure. Figure S2. Beta diversity PERMANOVA analysis for obesity, US exposure, and other co-variates, ranked by R2 Figure S3. Associations of gut microbiota and host obesity (BMI), stratified by: A. Sex; B. Age group (< 55 and > = 55 years); C. Place of birth (US born and non-US born); D. Hispanic origin (Caribbean [Dominican, Cuban, Puerto Rican] and Mainland [Central American, Mexican, South American]) ; E to J. Field Center (Bronx, Chicago, Miami, San Diego). Figure S4. Associations of gut microbiota and US exposure, further stratified by : A. Sex; B. Hispanic origin (Caribbean [Dominican, Cuban, Puerto Rican] and Mainland [Central American, Mexican, South American]); C. Obesity; D. Age group (< 55 and > = 55 years); E to J. Field Center (Bronx, Chicago, Miami, San Diego). Figure S5. Correlation heatmap for the 23 identified bacterial genera associated with both obesity and US exposure. Figure S6. Associations of 23 gut microbial genera with BMI, Body fat percentage and waist circumference. Figure S7. Correlation heatmap for all bacterial genera, using log ratios data and Clostridium as reference. Figure S8. Heatmap for overall dietary quality and individual dietary factor associated with gut microbial genera. Table S1. Characteristics of the study population by US exposure Table S3. Multivariable regression analyses: 23 genera associated with both obesity and US exposure. Table S10. Association of host obesity and US exposure with gut microbiota: species level analyses. Table S11. Conditional analysis for genera associated with both obesity and US exposure Table S12. Levels of gut microbial genera according to host obesity. Table S13. Target US exposure associated enzyme selection: Enrichment test for the annotated enzymes Table S14. Regression analyses: Enrichment test selected key enzymes associated with both US exposure and fiber intake. Table S15. The presence of key enzymes in specific bacteria: Alignment analysis.
Additional file 2: Table S2. Associations of 84 gut bacterial genera with obesity and US exposure.
Additional file 3: Table S4. Associations of 84 gut bacterial genera with obesity and US exposure after further adjusted for Hispanic background.
Additional file 4: Table S5. Associations of 84 gut bacterial genera with US exposure and all covariates in the multivariable regression model.
Additional file 5: Table S6. Associations of gut bacterial genera with obesity (BMI), stratified by Hispanic background groups.
Additional file 6: Table S7. Associations of gut bacterial genera with US exposure, stratified by Hispanic background groups.
Additional file 7: Table S8. Associations of gut bacterial genera with obesity (BMI), stratified by birth place, among the first generation immigrants.
Additional file 8: Table S9. Associations of gut bacterial genera with US exposure, stratified by birth place, among the first generation immigrants.
Data Availability Statement
HCHS/SOL data are archived at the National Institutes of Health repositories dbGap (accession number phs000810.v1.p1) and BIOLINCC (accession number HLB01141418a), and gut microbiome sequence data in this study are deposited in QIITA, ID 11666, and EMBL-EBI ENA, ERP117287 [65]. HCHS/SOL has established a process for the scientific community to apply for access to participant data and materials, with such requests reviewed by the project’s Steering Committee. These policies are described at https://sites.cscc.unc.edu/hchs/ (accessioned November 19, 2021). The corresponding author will accept reasonable requests for data and specimen access, which will be referred to the Steering Committee of the HCHS/SOL project.