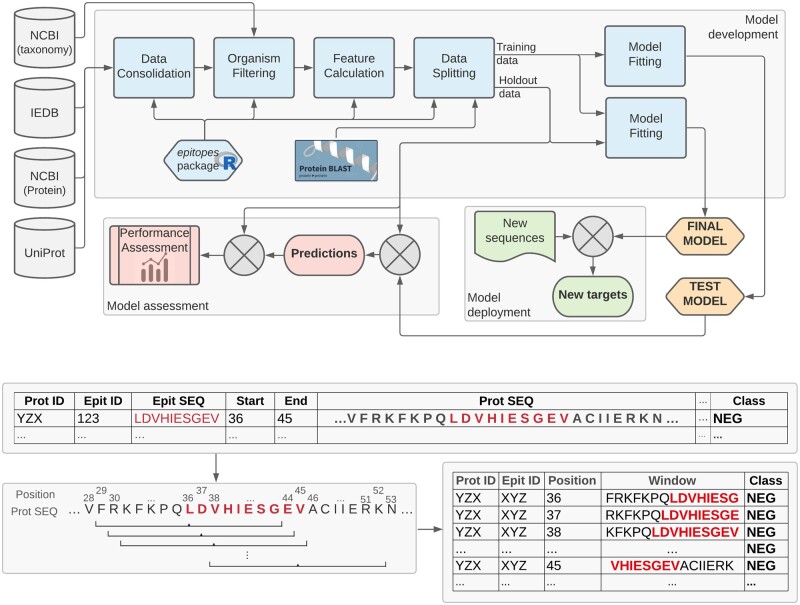
Fig. 1.
Top: Organism-specific epitope prediction pipeline. Publicly available data is retrieved from IEDB (Vita et al., 2019), NCBI (NCBI Resource Coordinators, 2015) and UniProt (UniProt Consortium, 2020) to compose an organism-specific dataset. 845 simple features are calculated for each AA, based on the local neighbourhood of every position extracted using a 15-AA sliding window representation with a step size of one (bottom). The data is then split at the protein level, based on protein ID and similarity, into a training set (used for model development) and a hold-out set (used to estimate the generalization performance of the models). The epitopes R package, which implements the main elements of this pipeline, is available at https://fcampelo.github.io/epitopes