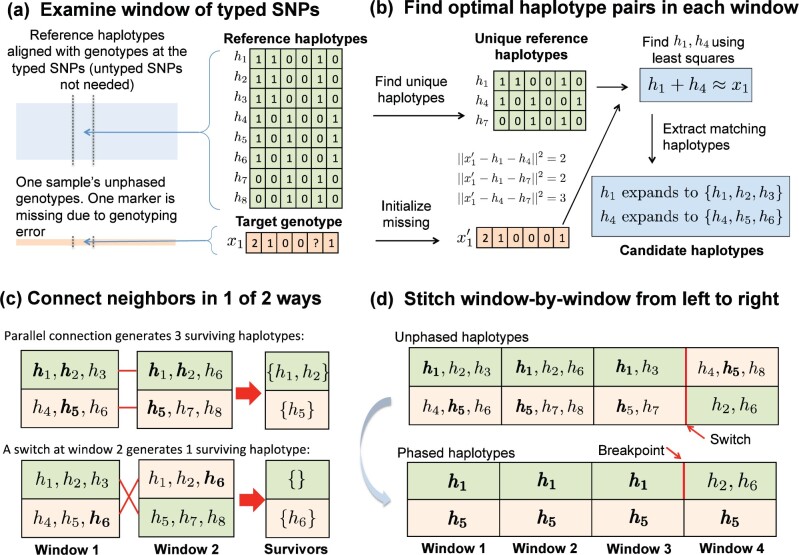
Fig. 1.
Overview of MendelImpute’s algorithm. (a) After alignment, imputation and phasing are carried out on short, nonoverlapping windows of the typed SNPs. (b) Based on a least squares criterion, we find two unique haplotypes whose vector sum approximates the genotype vector on the current window. Once this is done, all reference haplotypes corresponding to these two unique haplotypes are assembled into two sets of candidate haplotypes. (c) We intersect candidate haplotype sets window by window, carrying along the surviving set and switching orientations if that result generates more surviving haplotypes. (d) After three windows, the top extended chromosome possesses no surviving haplotypes, but a switch to the second orientation in the current window allows to survive on the top chromosome. Eventually, we must search for a break point separating from or between windows 3 and 4 (bottom panel).