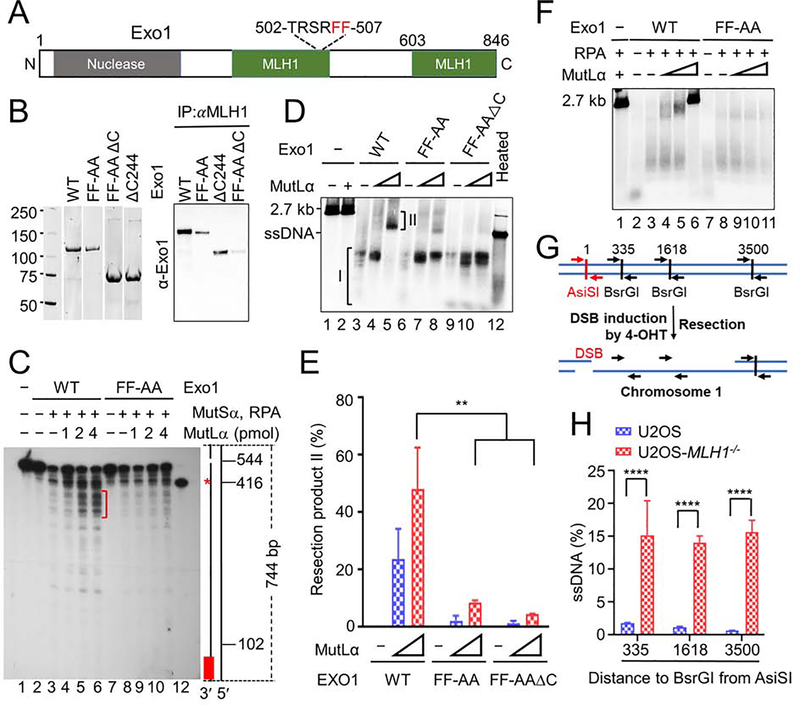
Figure 3. MutLα regulates Exo1 nuclease activity.
(A) Diagram of major functional domains in Exo1. (B) Co-immunoprecipitation–Western analysis of MutLα interactions with WT and mutant Exo1 (right) using purified proteins (left). (C) Southern blot analysis determining the impact of the MutLα–Exo1 interaction on mismatch-provoked excision in a purified MMR system. The excision products were digested with SspI and processed for Southern blot analysis, as described in the Methods section. Schematic representation of the 5′ G-T heteroduplex after SspI digestion is shown on the right side of the gel. Positions of the nick and mismatch (red asterisk) are 544-bp and 416-bp away, respectively, from the bottom SspI site. Red bar indicates the 32P-labeled oligonucleotide probe, which is complementary to the nicked strand near the bottom SspI site. Red bracket shows mismatch-provoked excision products terminated upon mismatch removal in reactions with WT Exo1, but in those with Exo1-FF-AA. (D) In vitro end resection assay to determine the impact of the MutLα–Exo1 interaction on Exo1-catalyzed resection using purified proteins and a linearized 2.7-kb pUC19 plasmid DNA. (E) Percentage of end resection product II shown in D in three independent assays. (F) In vitro end resection assay to determine the role of RPA in Exo1-catalyzed resection. (G) Principle of in vivo end resection assay. (H) qPCR analysis determining the amount of ssDNA generated at a specific DBS site (AsiSI) in WT and MLH1−/− U2OS cells. Data represent the mean ± SEM of 3 independent experiments (E) or 3 replicates (H). P value was calculated using one-way ANOVA. **, p<0.01; ***, p<0.001; and ****, p<0.0001.