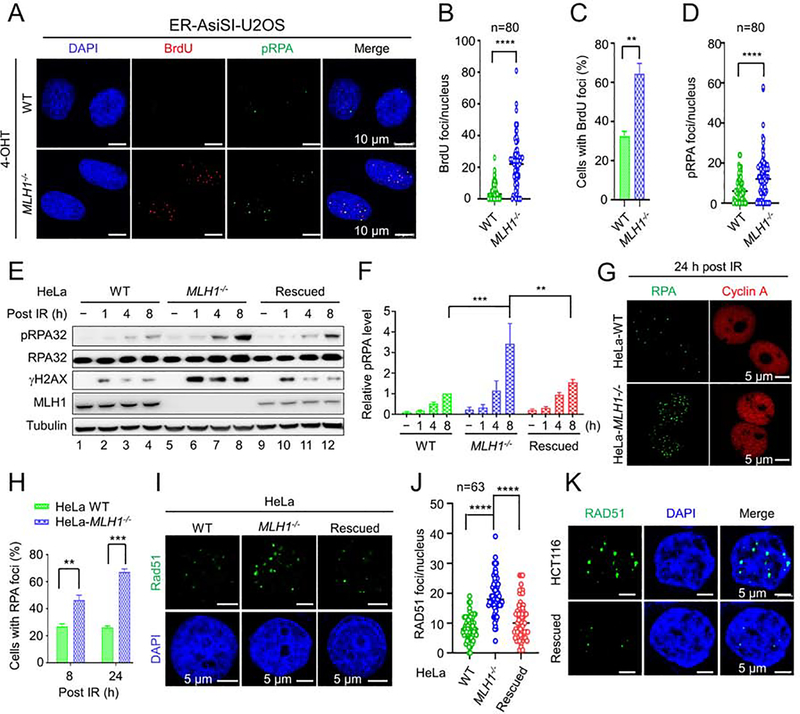
Figure 5. RPA exhaustion and aberrant resection intermediates in MLH1−/− cells.
(A) Microscope imaging showing BrdU incorporation by DNA polymerase using hyperesection-generated unprotected RPA as a template for DNA synthesis in the RPA exhaustion assay. ssDNA binding by phosphorylated RPA (pRPA) is also shown. (B) and (C) Quantification of BrdU foci/cell and percentage of cells exhibiting BrdU foci, respectively, in WT and MLH1−/− U2OS cells. (D) Quantification of pRPA foci per cell. (E) Western blots detecting pRPA and its association with DNA break marker γH2AX in the indicated cells before and after IR. (F) Quantification of relative pRPA levels shown in E, with 3 independent assays. (G) Immunoflurescence confocal analysis showing large RPA foci in HeLa MLH1−/− cells. (H) Quantification and comparison of the percentage of WT and MLH1−/− cells displaying RPA foci. (I) Immunofluorescence confocal analysis showing large Rad51 foci in MLH1−/− HeLa cells. (J) Quantification of RAD51 foci/nucleus in various HeLa cells, as indicated. (K) Immunofluorescence confocal analysis showing large Rad51 foci in HCT116 and MLH1-rescued HCT116 cells. Data represent the mean ± SEM of 3 independent experiments (C, F and H) or the indicated number of cells (B, D and J). P value was calculated using one-way ANOVA. **, p<0.01; ***, p<0.001; and ****, p<0.0001.