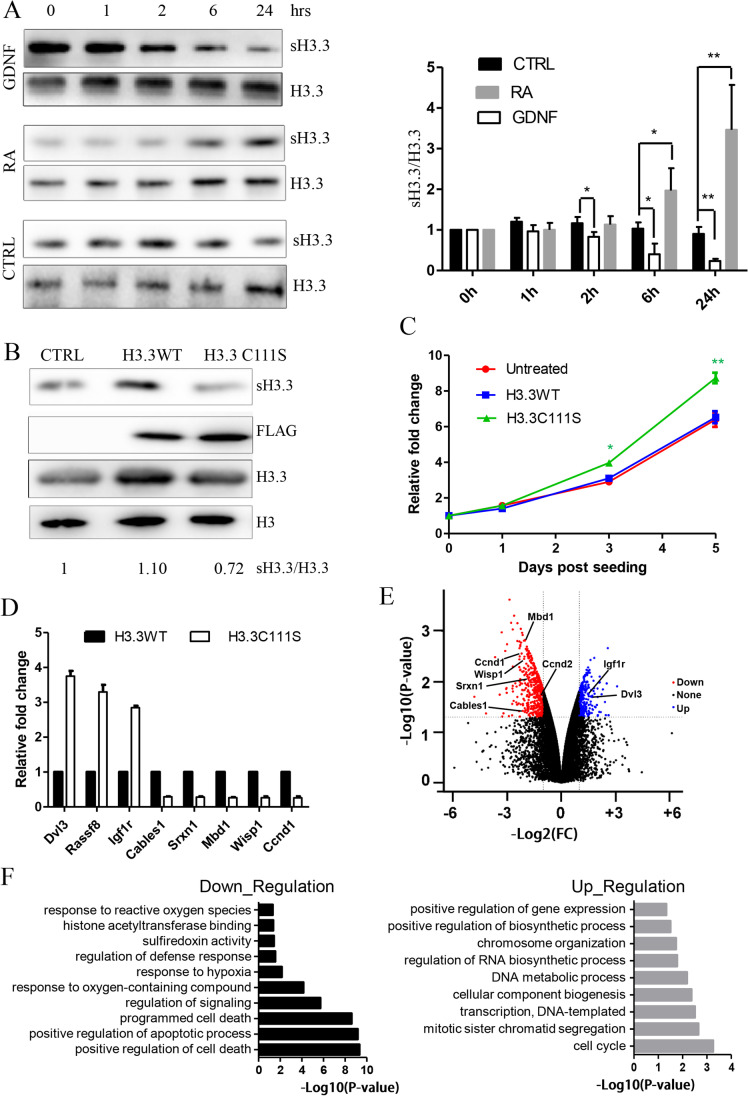
Fig. 5.
Effect of ectopic expression of the mutant H3.3 with C111S on cell growth rate and gene expression in the C18-4 cells. (A) Effects of GDNF and RA on sH3.3 expression in the C18-4 cells. Twenty-four hours after plating, the C18-4 cells were added with GDNF (50 ng/ml) and RA (1.0μM), respectively. The cells were harvested for measurement of sH3.3 expression at the indicated times after the treatment. Error bar denotes mean ± SEM. *P < 0.05 and **P < 0.01. (B) Expression of sH3.3 and H3.3 in the C18-4 cells infected with the recombinant Lentivirus that expressed wild-type and mutant (C111S) H3.3. The cultured C18-4 cells were harvested for the analysis when their confluence reached approximately 80–90%. These results represented one of three independent experiments with the similar data. (C) The growth rates of C18-4 cells. Untreated C18-4 cells and infected C18-4 cells which overexpressed wild-type or mutant H3.3 were seeded as described in “Experimental procedures,” and harvested for the counting of cell number at the indicated times after seeding. (D) Volcano plot showing differential expression of protein-coding genes between mutated and normal samples. Red and blue dots indicate significantly down-regulated (p<0.05 and log2FC<-1) and significantly up-regulated (p<0.05 and log2FC>1) differential expression, respectively. (E) RNA-seq data validation by quantitative RT-PCR. Expression of select up-regulated and down-regulated genes from the RNA-seq analysis was measured by quantitative RT-PCR in the C18-4 cells. (F) Biologic processes that are enriched in genes down-regulated (left) and up-regulated (right).