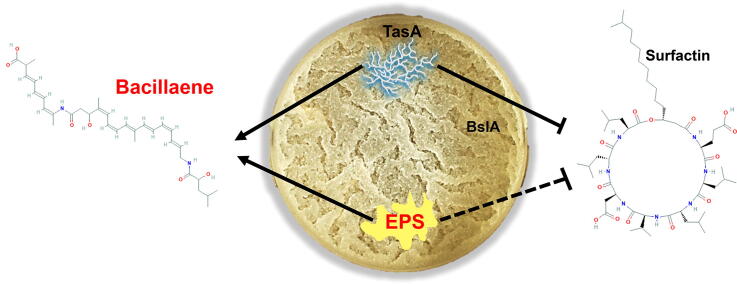
Graphical abstract
Keywords: Biofilms, Extracellular matrix, Imaging flow cytometry, Gene regulation
Abbreviations: CM, Conditioned medium; ECM, Extracellular matrix; LC-MS, Liquid chromatography–mass spectrometry; NRPs, Nonribosomal peptides; PKS, Polyketides; WT, Wild type
Abstract
In nature, bacteria frequently reside in differentiated communities or biofilms. These multicellular communities are held together by self-produced polymers that allow the community members to adhere to the surface as well as to neighbor bacteria. Here, we report that exopolysaccharides prevent Bacillus subtilis from co-aggregating with a distantly related bacterium Bacillus mycoides, while maintaining their role in promoting self-adhesion and co-adhesion with phylogenetically related bacterium, Bacillus atrophaeus. The defensive role of the exopolysaccharides is due to the specific regulation of bacillaene. Single cell analysis of biofilm and free-living bacterial cells using imaging flow cytometry confirmed a specific role for the exopolysaccharides in microbial competition repelling B. mycoides. Unlike exopolysaccharides, the matrix protein TasA induced bacillaene but inhibited the expression of the biosynthetic clusters for surfactin, and therefore its overall effect on microbial competition during floating biofilm formation was neutral. Thus, the exopolysaccharides provide a dual fitness advantage for biofilm-forming cells, as it acts to promote co-aggregation of related species, as well as, a secreted cue for chemical interference with non-compatible partners. These results experimentally demonstrate a general assembly principle of complex communities and provides an appealing explanation for how closely related species are favored during community assembly. Furthermore, the differential regulation of surfactin and bacillaene by the extracellular matrix may explain the spatio-temporal gradients of antibiotic production within biofilms.
1. Introduction
In terrestrial microenvironments, microbial communities often provide beneficial effects to other organisms, e.g., biocontrol agents grown on the surface of plant roots, thereby preventing the growth of bacterial and fungal pathogens [1].This beneficial effect is often associated with the production of antimicrobial substances and antibiotics [1]. Antibiotic production was shown to be regulated as well as affected by the biological, chemical and physical features of the environment [2], [3], [4]. However, the guiding rule for coordinating antibiotic production with the biotic environment remains unknown.
In many natural scenarios, bacteria grow in heterogeneous communities organized into a complex 3D structure, designated as biofilms [5]. The 3D structure of the biofilm was suggested to relieve metabolic stress, by the utilization of channels formed below the ridges and wrinkles within the colony that may facilitate diffusion of fluids, nutrients and oxygen [6], [7], [8], [9].
The formation of a biofilm is a developmental process, in which various genetic programs are activated in a specific order in different subpopulations of cells, for the proper establishment of a functional structure [7], [9], [10], [11], [12], [13], [14]. This apparent coordination can be explained by the temporally distinct exposure of cell subpopulations to specific microenvironments [13].
To form a functional structure, biofilm-forming cells produce polymers that constitute the extracellular matrix (ECM), where they bind to each other and to the surface. The ECM plays an important role in the resistance and resiliance of the entire biofilm community [15], [16], [17]. Although the ability to generate an ECM appears to be a common feature of multicellular bacterial communities, there is remarkable diversity in the means by which these matrices are constructed [18]. The most extensively studied components of biofilm ECMs are carbohydrate-rich polymers (i.e., extracellular polysaccharides or exopolysaccharides (EPS)), proteins, nucleic acids [18], [19] and biogenic minerals [20], [21].
Bacillus subtilis is a spore-forming facultative anaerobe and is associated with the rhizosphere and soil microbiomes [22]. This bacterium often serves as a model organism for beneficial Gram-positive bacteria, and its undomesticated strains form robust and architecturally complex biofilms [23].
B. subtilis is capable of forming various types of biofilms including on solid surfaces (colony), in the air–water interface (pellicles or floating biofilms) and submerged biofilms in domesticated strains [18]. The ECM of B. subtilis is characterized by the exopolysaccharides encoded by the epsA-O operon and the proteins TasA and BslA [22], [24]. TasA is a functional amyloid and is encoded by the tapA-sipW-tasA operon [24], [25], [26]. The biofilm surface layer protein A (BslA) is involved in the formation of a water-repellent hydrophobic coat over the biofilm and is critical for pellicle and colony biofilms [27], [28]. All matrix components were expressed and promoted during pellicle formation. Interestingly, all extracellular matrix components were shown to have additional non-structural roles: The protein TasA was shown to regulate motility, extracellular matrix production and the general stress response [29], [30] and exopolysaccharides were shown to activate the master regulator Spo0A [31] which regulates biofilm formation, sporulation and the production of the antibiotic Bacillaene [23], [32], [33]. Therefore, the role of the extracellular matrix in microbial competition could be multifactorial due to its independent structural and non-structural roles.
Here, we used imaging flow cytometry to evaluate the roles of all major extracellular matrix components in microbial competition for floating biofilm (pellicle) formation. The population within B. subtilis biofilms and root associated communities is heterogeneous [14], [34], [35], a heterogenicity which is affected by the production of the extracellular matrix. To overcome this fundamental property of biofilms, we monitored the effects of the extracellular matrix on the single cell level, relying on imaging flow cytometry. This method combines the power and speed of traditional flow cytometers with the resolution of the microscope. It therefore allows for high rate complex morphometric measurements in a phenotypically defined way [36], [37].
We uncovered that exopolysaccharides, but not the proteinous matrix components, have a dual role in excluding non-self-community members from mixed communities while promoting the self-aggregation and co-aggregation with related species. This role was indirect, and could be attributed to the exopolysaccharides dependent production of bacillaene within floating biofilms.
2. Results
2.1. Conditioned medium from B. subtilis pellicles is selectively toxic towards potential competitors
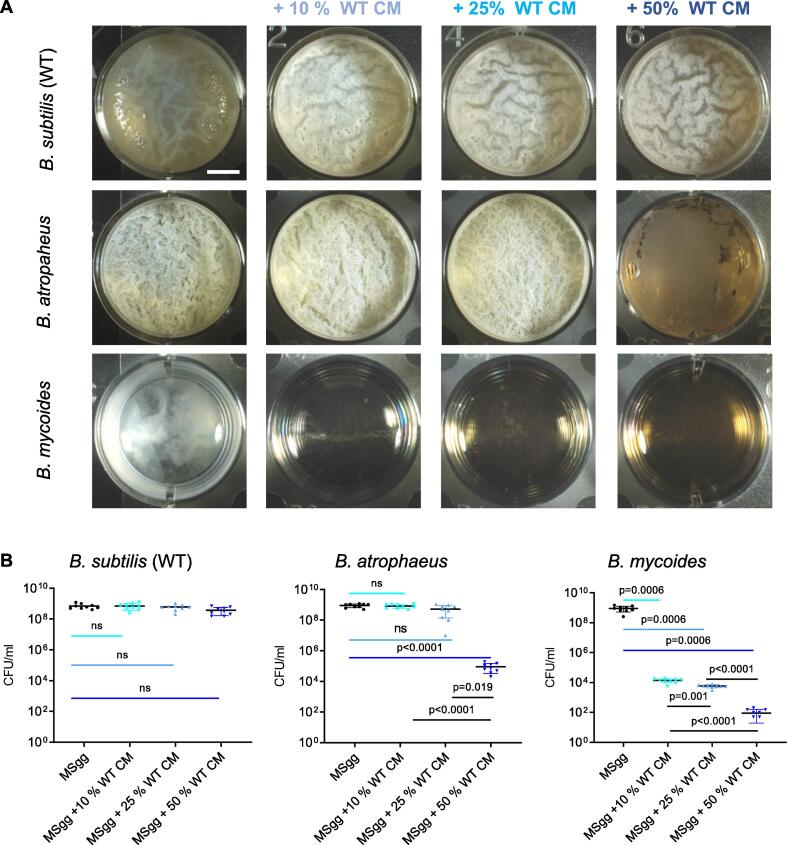
To test the effect of secreted products on the competition between related and non-related floating biofilms (pellicles), we collected the conditioned growth medium from B. subtilis pellicles and assessed its bioactivity towards three related Bacillus species: Bacillus subtilis (self), the highly related bacterium B. atrophaeus, and the phylogenetically distinct bacterium B. mycoides. B. subtilis clade members: B. subtilis and B. atrophaeus formed pellicles which were enhanced when supplemented with CM from B. subtilis (Fig. 1A). This result is consistent with previous findings that several secreted biofilm inducers: LuxS products (responsible for AI-2 pheromone production) [38], surfactants [1], [39] and quorum sensing peptides [40] were reported in the conditioned medium of these species. For B. mycoides, no pellicle formation was observed (control), however, growth was observed as cell aggregates were present in suspension. Both antagonistic and mutualistic effects on pellicle formation were evident within the first 24 h (Fig. S1), and lingered throughout pellicle maturation (Fig. 1).
Fig. 1.
Conditioned medium from B. subtilis pellicles is selectively toxic towards potential competitors. A) Monitoring pellicle formation and growth of the indicated strains when grown in MSgg medium and MSgg medium supplemented with conditioned medium (CM) from WT B. subtilis in increasing concentrations (10%, 25% and 50% v/v). Cells were grown at 30 °C and images were obtained at 72 h post inoculation and are representative of data presented in B. B) The number of CFU obtained when indicated strains were either grown in MSgg medium or MSgg medium supplemented with conditioned medium (CM) from WT B. subtilis in increasing concentrations (10%, 25% and 50% v/v). Cells were grown at 30 °C, and were harvested at 72 h post inoculation. Graphs represent mean ± SD from three independent experiments (n = 9). Statistical analysis was performed using Brown-Forsthye and Welch’s ANOVA, with Dunnett’s T3 multiple comparisons test. P < 0.05 was considered statistically significant. Scale bar = 5 mm.
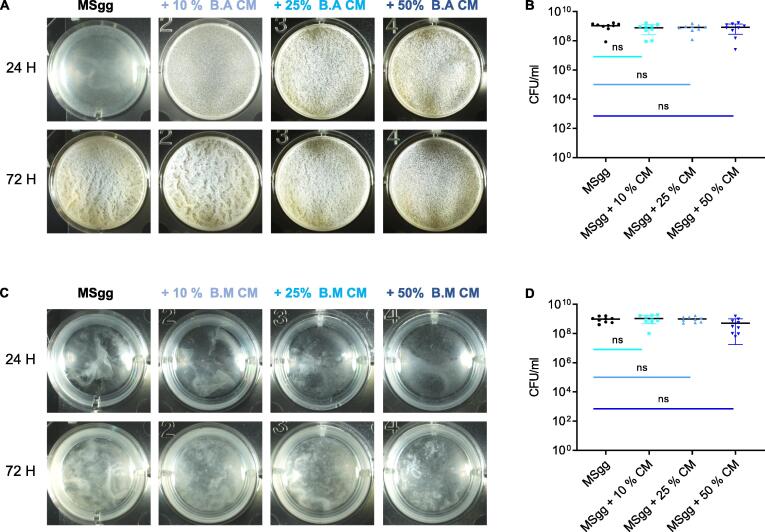
A dose-dependent response analysis of all examined species for pellicle formation confirmed a correlation between the relatedness of the strains and their response to the conditioned medium of B. subtilis (Fig. 1). B. subtilis was fully resistant to its own conditioned medium, B. atrophaeus was resistant to low concentrations of, but vulnerable to high concentrations of the B. subtilis conditioned medium, (50%v/v., p value < 0.0001) and B. mycoides was sensitive to all of the tested concentrations of the B. subtilis conditioned medium (p = 0.0006) (Fig. 1B). Furthermore, both B. atrophaeus and B. mycoides were resistant to their own conditioned medium, and no significant differences were observed on pellicle formation or growth (Fig. 2). These results are consistent with our previous work where we reported that the conditioned rich medium of B. subtilis pellicles was inert to self, slightly toxic to the related B. atrophaeus, and significantly toxic to B. mycoides [41].
Fig. 2.
B. atrophaeus and B. mycoides were resistant to their own conditioned medium. A) Monitoring pellicle formation and growth of the indicated strains when grown in MSgg medium and MSgg medium supplemented with conditioned medium (CM) from B. atrophaeus in increasing concentrations (10%,25% and 50% v/v). Notably, 25% CM induced a highly stable pellicle that did not change significantly over time. Cells were grown at 30 °C and images were obtained at 24 h and 72 h post inoculation and are representative of data presented in B. B) The number of CFU obtained when indicated strains were either grown in MSgg medium or MSgg medium supplemented with conditioned medium (CM) from B. atrophaeus in increasing concentrations (10%, 25% and 50% v/v). Cells were grown at 30 °C, and harvested at 72 h post inoculation. C) Monitoring pellicle formation and growth of the indicated strains when grown in MSgg medium and MSgg medium supplemented with conditioned medium (CM) from B. mycoides in increasing concentrations (10%, 25% and 50% v/v). Cells were grown at 30 °C and images were obtained at 24 h and 72 h post inoculation and are representative of data presented in D. D) The number of CFU obtained when indicated strains were either grown in MSgg medium or MSgg medium supplemented with conditioned medium (CM) from B. mycoides in increasing concentrations (10%, 25% and 50% v/v). Cells were grown at 30 °C, and were harvested at 72 h post inoculation. Graphs represent mean ± SD from three independent experiments (n = 9). Statistical analysis was performed using Brown-Forsthye and Welch’s ANOVA, with Dunnett’s T3 multiple comparisons test. P < 0.05 was considered statistically significant. Scale bar = 5 mm.
2.2. The exopolysaccharides positively regulate the selective toxicity of the conditioned medium
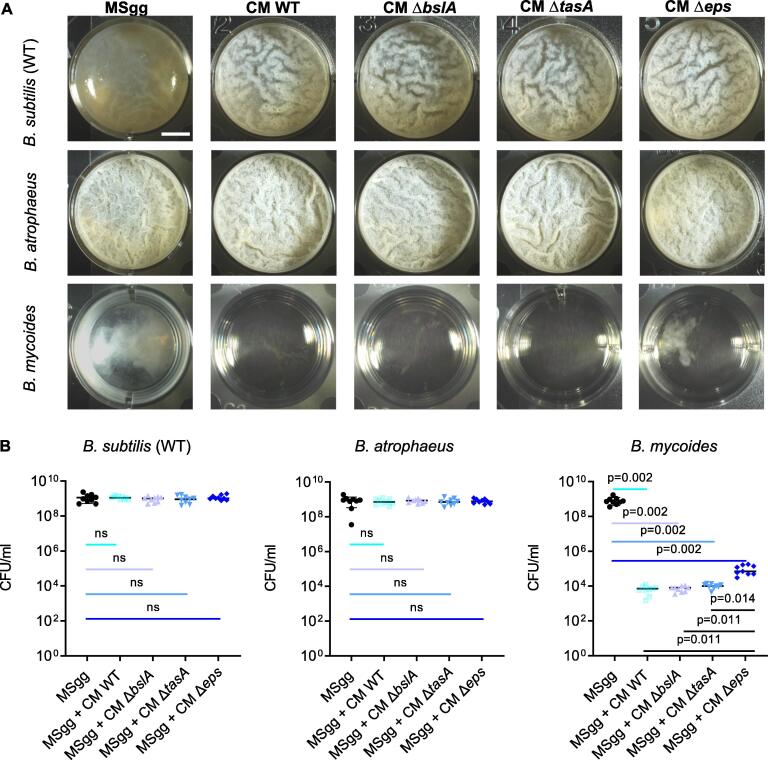
To compare the potential non-structural roles of the extracellular matrix components of B. subtilis on interspecies competition, we collected the conditioned media of all three ECM mutants (Fig. 3A) and compared its bioactivity to the conditioned media of the WT B. subtilis on recipient cultures. As shown, a deletion of the biosynthetic clusters of the exopolysaccharides (Δeps) significantly reduced the toxicity of the conditioned medium towards B. mycoides. Quantifying B. mycoides cells grown in the presence of conditioned medium from all donors, confirmed the specific effect of the deletion of the exopolysaccharides resulting in a significant rescue of B. mycoides. To test whether this non-structural role of exopolysaccharides was direct, we then supplemented the medium with increasing concentrations of either purified exopolysaccharides (eps) or TasA protein to study if they have any effect on the growth of B. mycoides. Interestingly, instead of having any inhibitory effect on the growth, these purified ECM components facilitated the growth of B. mycoides (Figs. S2,S3), potentially serving as carbon and nitrogen sources. This suggested that ECM components were not directly responsible for toxicity towards B. mycoides. Therefore, we hypothesized that the antagonistic effect of the extracellular matrix and the exopolysaccharides towards the phylogenetically distinct competitor B. mycoides is indirect.
Fig. 3.
The exopolysaccharides positively regulate the selective toxicity of the conditioned medium. A) Monitoring pellicle formation and growth of the indicated strains when grown in MSgg medium and MSgg medium supplemented with conditioned medium (CM) from WT B. subtilis and its ECM mutants (10%, v/v). Cells were grown at 30 °C and images were obtained at 72 h post inoculation and are representative of data presented in B. B) The number of CFU obtained when indicated strains were either grown in MSgg medium or MSgg medium supplemented with conditioned medium (CM) from WT B. subtilis and its ECM mutants (10%, v/v). Cells were grown at 30 °C, and were harvested at 72 h post inoculation. Graphs represent mean ± SD from three independent experiments (n = 9). Statistical analysis was performed using Brown-Forsthye and Welch’s ANOVA, with Dunnett’s T3 multiple comparisons test. P < 0.05 was considered statistically significant. Scale bar = 5 mm.
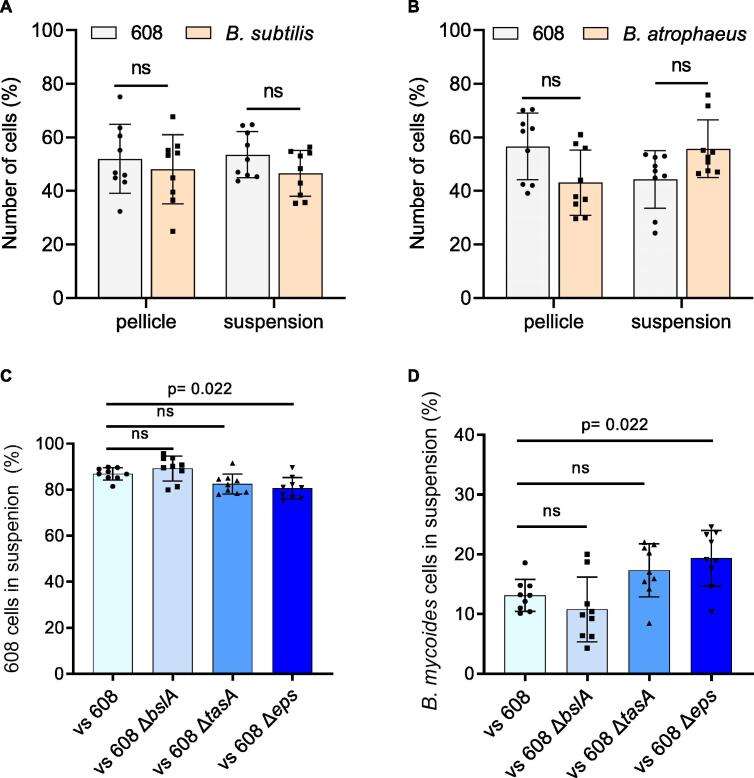
To assess the structural and non-structural roles of exopolysaccharides and proteinous matrix during interspecies competitions, we competed B. subtilis and its matrix mutants versus itself, B. atrophaeus (mostly resistant to the conditioned media) and B. mycoides (mostly sensitive to the conditioned media) for pellicle formation. Pellicle formation is a system which is less prone to diffusion limitations as compared with semi-solid agar surfaces, and therefore allows us to better quantify the response of the different competitors to secreted products. For strains capable of forming a pellicle, we compared the presence of the different competitors within the pellicle and within the suspension separately. To get accurate results we relied on imaging flow cytometry, a technique which allows us to image the cells and to fully distinguish between and accurately quantify constitutively expressing B. subtilis Phyperspank-gfp (608) or its ECM mutants vs their competitor’s: WT B. subtilis, B. atrophaeus and B. mycoides.
As shown in (Fig. 4A,B), B. subtilis was capable of forming pellicles in an exopolysaccharide-dependent manner with itself and with B. atrophaeus as well as to co-aggregate with these strains. During competition with B. mycoides no significant differences were observed in pellicle formation of B. subtilis WT and its ECM mutants regardless of the presence of the extracellular matrix, indicating a barrier for joint pellicle formation (Fig. S4). However, the presence of free living B. mycoides cells in suspension was detected and these free living cells co-existed significantly better with B. subtilis strain lacking the exopolysaccharide biosynthetic operon (Δeps) (p = 0.022) than with the WT B. subtilis (Fig. 4C, D).
Fig. 4.
Exopolysaccharides prevent growth of free-living cells of the sensitive competitor B. mycoides. Quantifying the number of cells present in the pellicle/suspension of each competition assay using imaging flow cytometry. A) A competition assay was set up between WT B. subtilis stains harboring Phyerspank-gfp (608), and WT B. subtilis in MSgg medium. Quantification of number of cells of each strain both in pellicle and suspension was performed using imaging flow cytometry. B) A competition assay was set up between WT B. subtilis stains harboring Phyerspank-gfp (608), and B. atrophaeus in MSgg medium. Quantification of number of cells of each strain both in pellicle and suspension was performed using imaging flow cytometry. C) A competition assay was set up between WT B. subtilis stains harboring Phyerspank-gfp (608), 608 ΔtasA, 608 ΔbslA and 608 Δeps mutants vs B. mycoides, in MSgg medium. From the suspension culture, number of 608 and its deletions mutants cells, in competition against B. mycoides cells were quantified using imaging flow cytometry. D) A competition assay was set up between WT B. subtilis stains harboring Phyerspank-gfp (608), 608 ΔtasA, 608 ΔbslA and 608 Δeps mutants vs B. mycoides, in MSgg medium. From the suspension culture, number of B. mycoides cells in competition against 608 and its deletion mutant cells were quantified using imaging flow cytometry. Data were collected 72 h post inoculation, and 100,000 cells were counted. Graphs represent mean ± SD from three independent experiments (n = 9). The data were analyzed using IDEAS 6.3. For A-B, statistical analysis was performed using two-way ANOVA followed by Dunnett’s multiple comparison test. For B-C, statistical analysis was performed Brown-Forsthye and Welch’s ANOVA, with Dunnett’s T3 multiple comparisons test. P < 0.05 was considered statistically significant.
2.3. Exopolysaccharides regulate the antibiotic bacillaene to eliminate sensitive competitors
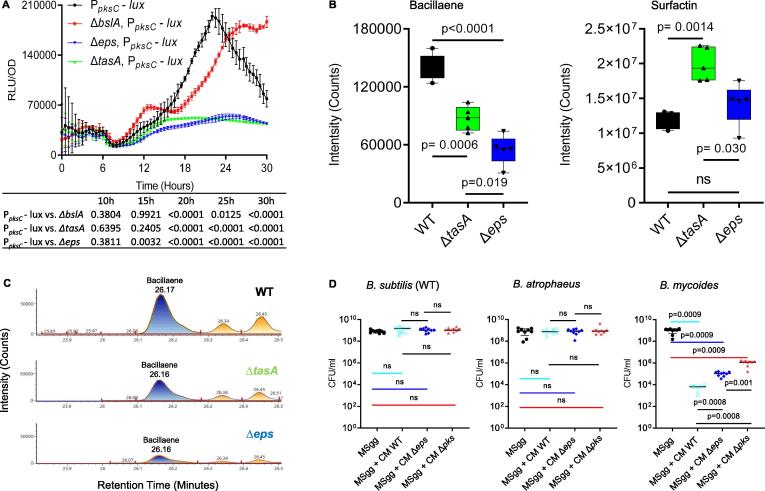
Recently, we found that two antibiotics: the NRP (non-ribosomal peptide) surfactin and the polyketide bacillaene play an important role in the elimination of phylogenetically distinct competitors over semi-solid surfaces and in shaking culture [41]. These results are consistent with our finding that B. subtilis and B. atrophaeus contains fully conserved NRPs/PKS gene clusters, while the same gene clusters are not present in B. mycoides (Maan et al., Nature Communications, In press). As we found that the role of exopolysaccharides (Δeps) during competition is largely indirect, and was pronounced in the free-living cells present in suspension, we tested whether the exopolysaccharides are capable to regulate surfactin and bacillaene production. The expression of operons encoding surfactin and bacillaene biosynthetic clusters tagged to a luciferase reporter [41] were compared between the parental strain (WT) and its ECM mutant derivatives. Notably, luciferase cannot be accumulated in bacterial cells [42], and therefore luciferase essays reflect the real-time expression of the antibiotics, both under the temporal regulation of several transcription factors [33], [43].
Both exopolysaccharides and TasA were required for optimal bacillaene expression (Fig. 5A); Consistently, LC-MS quantification of bacillaene production verified that it is induced by exopolysaccharides and repressed by TasA (Fig. 5B,C). Notably, a deletion of TasA induced the expression (Fig. S5) and production of surfactin (Fig. 5B). These results could explain why deletion of Δeps has a more pronounced effect on the toxicity towards B. mycoides, compared with ΔtasA. Neither purified exopolysaccharides nor TasA were sufficient to restore the expression from pks promoter (Fig. S6), indicating that these ECM components either act as a complex or that the nature of the signal is multilayered.
Fig. 5.
Exopolysaccharides and the antibiotic Bacillaene are essential to eliminate sensitive competitors. A) Analysis of the luciferase activity in a WT B. subtilis strain harboring PpksC-lux (bacillaene) reporter, and its indicated ECM deletion mutants. Luminescence was monitored in MSgg medium, at 30 °C for 30 h. Graphs represent mean ± SD from three independent experiments (n = 9). Statistical analysis was performed using two-way ANOVA followed by Dunnett’s multiple comparison test. P < 0.05 was considered statistically significant. P values at different time points are shown in the table. Luciferase activity was normalized to avoid artifacts related to differential cell numbers as RLU/OD. B) Liquid chromatography-mass spectrometry analysis of surfactin and bacillaene from WT B. subtilis and its ΔtasA and Δeps mutant derivatives grown in untreated MSgg medium to an OD ≈ 0.7. Supernatant was extracted from the samples using HCl treatment. Box and whisker plot shows median and interquartile range, together with the maximum and minimum values from five biological repeats (n = 5). C) Representative peaks from liquid chromatography-mass spectrometry analysis of bacillaene from the WT B. subtilis and its ΔtasA and Δeps mutants grown in MSgg medium. Peak area is correlative to the concentration of baceilaene. D) The number of CFU obtained when indicated strains were either grown in MSgg medium or MSgg medium supplemented with conditioned medium (CM) from WT B. subtilis and its Δeps and Δpks mutants (10%, v/v). Cells were grown at 30 °C, and were harvested at 72 h post inoculation. Graphs represent mean ± SD from three independent experiments (n = 9). Statistical analysis was performed using Brown-Forsthye and Welch’s ANOVA, with Dunnett’s T3 multiple comparisons test. P < 0.05 was considered statistically significant.
To test whether the deletion of bacillaene biosynthetic clusters indeed mimics the effect of deletion of the exopolysaccharides, we examined the effect of a pks mutant (deficient in bacillaene biosynthesis) on the bioactivity of the conditioned medium. Accordingly, the deletion of pks significantly reduced the toxicity of the conditioned medium towards B. mycoides (Fig. 5D, S7). A deletion of srf on top of pks had mostly eliminated the toxicity of B. subtilis towards B. mycoides aggregates, indicating that both matrix-regulated antibiotics interact under these conditions (Fig. S8).
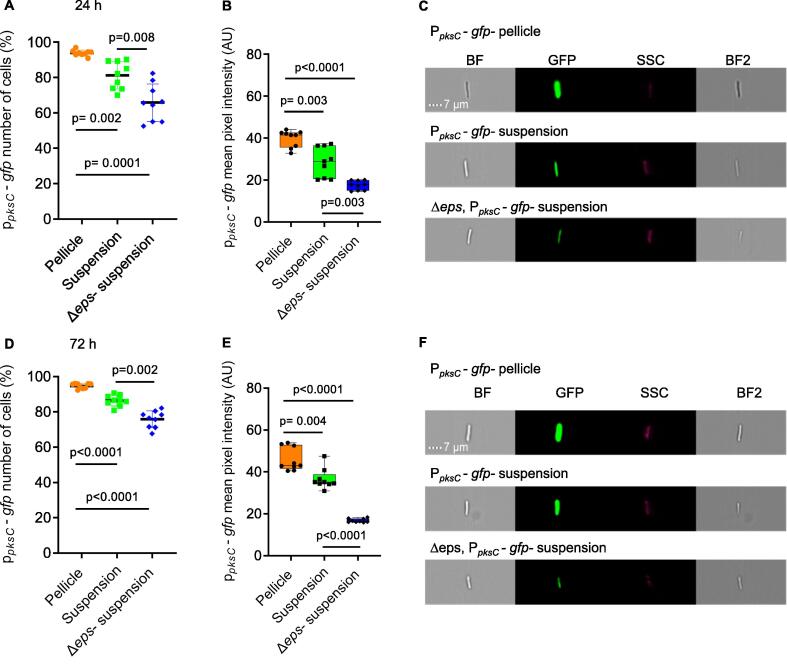
To confirm that matrix exopolysaccharides act independently of their structural role to induce bacillaene, we quantified the expression from the promoter of pks tagged to a GFP reporter using imaging flow cytometry in the parental strains, and its eps mutant. As shown (Fig. 6) the intensity of the expression from pks promoter and the amount of positively expressed cells were significantly reduced in the exopolysaccharides mutant (Δeps) throughout static growth. For WT B. subtilis cells growing in suspension, a reduced expression was observed as compared to the cells growing within the pellicle, however, cells growing in suspension also demonstrated a higher expression from pks promoter compared with the exopolysaccharides mutant. These results confirm a non-structural role for exopolysaccharides in inducing bacillaene, which contributes to the favorable outcome of interspecies interactions in biofilms.
Fig. 6.
Exopolysaccharides induce bacillaene expression. Monitoring the expression of bacillaene in WT B. subtilis strain and its Δeps mutant harboring Ppks-gfp reporter. The bacillaene expression was measured using imaging flow cytometry in both pellicle and suspension in for WT B. subtilis Ppks-gfp, and only in suspension for Δeps, B. subtilis Ppks-gfp mutant, due to its inability to form pellicle. A) Showing positively expressing fluorescent populations (%) at 24 h. B) Mean pixel intensity of the fluorescent positive populations as in A. C) Representative bright-field and fluorescent images related to expression of reporters in A, scale bar = 7 µm. D) Showing positively expressing fluorescent populations (%) at 72 h. E) Mean pixel intensity of the fluorescent positive populations as in A. F) Representative bright-field and fluorescent images related to expression of reporters in F, scale bar = 7 µm. Graphs A-D represent mean ± SD from three independent experiments (n = 9). Graphs B-E represent box and whiskers plots showing median and interquartile range together with maximum and minimum values and outlier points from three independent experiments (n = 9). Statistical analysis was performed using Brown-Forsthye and Welch’s ANOVA, with Dunnett’s T3 multiple comparisons test. P < 0.05 was considered statistically significant.
3. Discussion
Bacteria in nature are most often found in the form of multicellular aggregates commonly referred to as biofilms [44], [45]. When compared to the planktonic (free-living) state, cells in biofilms are more protected from environmental insults, including sterilizing agents, antibiotics, and the immune system. Biofilms enable bacteria to attach more firmly to their hosts and better access to nutrients [46], [47], [48], [49], [50], [51], [52].
The self-produced extracellular matrix (ECM) surrounds and protects the cells, and makes them adhere to each other or to a surface [19]. This feature makes bacterial biofilms an especially appealing system in which to study multicellular development, with exopolysaccharides, carbohydrate rich polymers being the most studied and wide spread ECM component [18], [53].
Various genetic analyzes have provided strong evidence that biofilm exopolysaccharides play a fundamental structural role in different bacterial species, impact bacterial virulence, and promote capsule formation [54], [55], [56], [57], [58], [59]. The biofilm of B. subtilis contains several exopolysaccharide polymers, produced by the epsA-O operon, and composed of glucose, galactose and N-acetyl-galactosamine [60], [61]. Colonies of mutants in the epsA-O operon, and specifically the glycosyltransferase gene epsH, lack the exopolysaccharide component of the ECM and are featureless, as opposed to the wrinkled wild type colony [23].
Here we report that in addition to their structural role, manifested by their necessity for B. subtilis to generate pellicles (floating biofilms) with itself [62] and with a related bacterium, B. atrophaeus (Fig. 1), the exopolysaccharides act to induce the production of bacillaene, a polyketide antibiotic [33], [63], [64], [65] which was recently shown to be essential for the elimination of phylogenetically distinct Bacillus species [41], [66]. Our results indicate that this non-structural role of the exopolysaccharides is manifested by a capacity of B. subtilis to repel the phylogenetically distinct competitor, B. mycoides from the pellicle and its surrounding media.
Interestingly, the protein matrix component TasA induced Bacillaene while repressing the production of the surfactant and antibiotic surfactin [67], [68], also involved in horizontal gene transfer [69]. This result may explain the heterogeneity in gene expression of antibiotics within the biofilm [41], and predicts that their local expression may also reflect the local concentration of the extracellular matrix components.
So far we could observe two global signals for inducing antibiotic production in B. subtilis: peptidoglycan [41] and plant secretions [37] that induced both surfactin and bacillaene. In contrast, ECM driven regulation of antibiotic production is highly specific: TasA represses surfactin and induces bacillaene, and exopolysaccharides trigger the production of bacillaene but to some extent represses surfactin expression, indicating an overall fine-tuning of antibiotic production. One advantage of differential ECM-driven regulation is a potential capacity to control local expression within the complicated biofilm structure [18], [70].
When considering a developmental model for biofilm formation, it is tempting to speculate that the bacterial ECM is involved in regulation of genetic programs in designated subpopulation of cells in the biofilm [18]. It has been evident that in multicellular eukaryotes, the production of communication factors depends on cell–ECM interactions [71]. In this work, we describe a similar role for the exopolysaccharides in changing the decision-making processes and the competitiveness of the bacterial biofilm-forming cells. Furthermore, we demonstrate how this ECM-derived cue maintains an essential subset of antibiotic producing cells within the biofilm population.
4. Materials and methods
4.1. Strains and media
All strains used in this study are listed in Supplementary Table 2.
For cloning purposes, selective media was prepared using LB broth or LB-agar using appropriate antibiotics at the following concentrations, 10 μg/ml chloramphenicol (Amersco), 10 µg/ml, 10 μg/ml spectinomycin (Tivan biotech), 10 µg/ml kanamycin (A.G scientific), 1 µg/ml erythromycin (Amresco) + 25 µg/ml lincomycin (Sigma-Aldrich) .MSgg was prepared as described previously [23]. Briefly, 5 mM potassium phosphate, 100 mM MOPS (pH 7), 2 mM MgCl2, 50 μM MnCl2, 50 μM FeCl3, 700 μM CaCl2, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, Threonine (50 μg ml−1), Tryptophan (50 μg ml−1), and PhenylAlanine (50 μg ml−1).
4.2. Strains construction
All strains used in this study are reported in Supplementary Table 2. Briefly, DNA was extracted from the indicated deletion strains using Wizard Genomic DNA Purification Kit (Promega) and transformed into the genome of indicated B. subtilis reporter strains induced for natural competence [72]. All mutant strains were confirmed for an accurate integration by PCR.
4.3. Conditioned medium from pellicle culture preparation
Conditioned medium (CM) refers to cell free extracts of the culture media: Cells from indicated strains were grown to a mid-logarithmic phase of growth and were diluted 1:1000 and suspended in MSgg medium to allow pellicle formation and growth. Cells were incubated at 30 °C (Brunswick™ Innova® 42) and grown to an OD of ≈ 0.7 in dark. As the ΔtasA and Δeps mutants grow faster than WT B. subtilis, it is important that cells are grown to the similar OD. After incubation, were removed by a centrifugation and CM was filtered using a 0.22 μm filter (Corning). For each independent experiment, a fresh CM was collected.
4.4. Pellicle assays
Cells were grown from a single colony isolated from LB plates, transferred to LB broth, and grown to a mid-logarithmic phase in 2-ml of LB (∼3 h at 37 °C). Cells were added to the 2 mL liquid MSgg in 24 well plates (Thermo Scientific). The cultures were then grown at 30 °C for 72 h in the dark. The pellicles were imaged at 24 h and 72 h. Cells were either grown in the presence or absence of CM as indicated in each corresponding figure legends. The pellicles were imaged using stereomicroscope (Zeiss), using Objective Plain 0.5 × FWD 134 mm lens at 10× magnification. Captured images were processed using Zen software (Zeiss).
For CFU count, pellicles were harvested at 24 h or 72 h by scrapping off the entire pellicle plus pipetting the entire underlying cell suspension, and suspending in PBS (phosphate-buffered saline). To separate the cells a BRANSON digital sonicator, was used at an amplitude of 10% with pulse of 5 s. This sonicated cells suspension was transferred to a Griener 90 well plate (Sigma-Aldrich) and a serial dilution ranging from 10−1 to 10−7 was performed in PBS. From these dilutions 20 µl of samples was transferred on a LB plate. Plates were incubated at 30 °C overnight and CFU were counted next day.
4.5. Growth measurements
Cells were grown from a single colony isolated from LB plates, transferred to LB broth, and grown to a mid-logarithmic phase. Cells were then grown in 300 μl of MSgg medium and MSgg medium supplemented with purified TasA and exopolysaccharides fractions in a 96-well microplate (Thermo Scientific), with agitation, at 30 °C for 30 h, in a microplate reader (Synergy 2; BioTek, Winooski, VT, USA). The optical density at 600 nm was measured every 30 min.
4.6. Luminescence analysis
Strains carrying the indicated luminescence reporters were grown from a single colony isolated over LB plates to a mid-logarithmic phase of growth. Cells were then grown in 300 μl MSgg medium, in a 96 well plate with white opaque walls and clear tissue culture treated flat bottoms (Corning). Measurements were performed every 30 min at 30 °C for 30 h, using a microplate reader (Synergy 2; BioTek, Winooski, VT, USA). Luciferase activity was calculated as RLU/OD, to avoid artefacts related to the luminescence intensity influenced by the bacterial growth rate.
4.7. TasA protein
TasA was expressed and purified as described in [30] and was a kind gift from the Romero lab.
4.8. EPS extraction
EPS was extracted from B. subtilis pellicles grown for 72 h at 30 °C in MSgg medium, as described in pellicle assays. In total 15 pellicle colonies were scrapped and suspended in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4). The pellicles were dissolved using sonication, the suspensions were centrifuged to remove the cells. The supernatant was then treated with five volumes of ice-cold isopropanol and incubated overnight at 4 °C. Samples were then centrifuged at 4 °C for 10 min at 7,500 × g. The pellets were collected and suspended in a digestion mix (0.1 M MgCl2, 0.1 mg/ml of DNase, and 0.1 mg/ml of RNase), and were incubated at 37 °C for 4 h. Following incubation samples were extracted twice using phenol–chloroform. The aquatic fraction obtained from phenol-chloroform was dialyzed using Slide-A-Lyzer dialysis cassettes (Thermo Fisher) for 48 h. A 3,500 molecular weight cut-off, against distilled (d)H2O was used. The obtained samples were then were lyophilized and final lyophilized fraction was dissolved in 500 μl of dH2O. The fractions were stored for further use at −80 °C.
4.9. Imaging flow cytometry
Indicated strains from the corresponding figure legends were grown in MSgg medium as described in pellicle assays. For flow cytometry analysis pellicle from the pellicle forming strains was gently scrapped off and separated from its suspension and pellicles were suspended in PBS. Both the pellicles and their suspension counterparts were then sonicated using a BRANSON digital sonicator, at an amplitude of 10% and a pulse of 5 sec to separate the cells without compromising their viability. These sonicated samples were then subjected to Imaging Flow Cytometry.
Data was acquired by ImageStreamX Mark II (AMNIS, part of Luminex corp., Austin Tx) using a 60X lens (NA = 0.9). GFP lasers were excited using 488 nm (200mW), and 785 nm (5mW) for side scatter measurement. Acquired bacterial cells were gated according to their area (in square microns) and side scatter. Wild type B. subtilis was used as a negative control, to separate GFP negative from true GFP positive cells. During acquisition, samples were run alongside calibration beads and calibration beads were excluded from the gate. Every bacterium was represented as a single event that was selected based to their area (in square microns) and aspect ratio (width divided by the length of a best-fit ellipse). Focused events were selected by the Gradient RMS and Contrast features. Contrast features were defined as measure of sharpness quality of an image by detecting large changes of pixel values in the image. To quantify GFP expression Mean Pixel feature (the mean of the background-subtracted pixels contained in the input mask) was utilized Intensity of GFP positive cells was defined by the sum of the background subtracted pixel values within the image, and by Max Pixel values (the largest value of the background-subtracted pixels) of the GFP channel (Ch02). For each sample, 100,000 events were acquired and analysed using IDEAS 6.3 (AMNIS).
4.10. Semipolar metabolite extraction and sample preparation
The dried extract was prepared as described in [73] with minor alterations. Briefly, B. subtilis and its ECM mutants were grown to a mid-logarithmic phase of growth. Cells were diluted 1:100 in 300 mL of in MSgg medium. Cells were grown at 30 °C in dark to an OD of 0.7 in a shaker incubator (Brunswick™ Innova® 42). Conditioned medium was obtained from the cultures as described previously. The pH of conditioned medium was then adjusted to 2.0 by addition of 6 mol/L HCl, and was kept overnight at 4 °C. The precipitated material from HCl treated conditioned medium were obtained by centrifugation at 10,000 × g for 20 min at 4 °C. The precipitates were dissolved in 0.5 mL methanol. The tubes were vortexed for half a minute, then sonicated for 30 min in an ice-cold sonication bath (taken for a brief vortex every 10 min), vortexed again, and then centrifuged at 13,000 rpm at 4˚C for 10 min. 200 µl supernatant was transferred into the HPLC vials for analysis.
4.11. LC-MS for semipolar metabolites analysis
Metabolic analysis of semipolar phase was performed using Waters ACQUITY UPLC system coupled to a Vion IMS QTof mass spectrometer (Waters Corp., MA, USA). The chromatographic separation was performed on an ACQUITY UPLC BEH C18 column (2.1 × 100 mm, i.d., 1.7 μm) (Waters Corp., MA, USA). The mobile phase A consisted of 95% water (UPLC grade) and 5% acetonitrile, with 0.1% formic acid; mobile phase B consisted of 100% acetonitrile with 0.1% formic acid. The column was maintained at 45˚C, and the flow rate of mobile phase was 0.4 mL*min−1. Mobile phase A was initially run at 100%, and it was gradually reduced to 72% at 22 min, following a decrease to 0% at 36 min. Then, mobile phase B was run at 100% until 38 min; then, mobile phase A was set to 100% at 38.5 min. Finally, column was equilibrated at 100% mobile phase A until 40 min. MS parameters were as follows: the source and de-solvation temperatures were maintained at 120 °C and 350 °C, respectively. The capillary voltage was set to 1 kV; cone voltage was set for 40 V. Nitrogen was used as de-solvation gas and cone gas at the flow rate of 700 L*h−1 and 50 L*h−1, respectively. The mass spectrometer was operated in full scan HDMSE positive ionization, over a mass range of 50–2000 Da. For the high energy scan function, a collision energy ramp of 20–70 eV was applied, for the low energy scan function 5 eV was applied. Leucine-enkephalin was used as lock-mass reference standard. Bacillaene was putatively identified using adducts, fragmentation, and mass accuracy. (See Supplementary Table 1).
4.12. Semipolar compounds identification and data analysis
LC-MS data were analyzed and processed with UNIFI (Version 1.9.4, Waters Corp., MA, USA). The putative identification of the Surfactin was performed by comparison of standard’s and sample’s accurate masses, fragmentation pattern, retention time, and ion mobility (CCS) values. The putative identification of the Bacillaene was performed by studying sample’s peak accurate masses, theoretical fragmentation pattern, retention time, and ion mobility (CCS) values (See Supplementary Table 1).
4.13. Statistical analysis
All experiments were performed three separate and independent times in triplicates, unless mentioned otherwise. Datasets were compared using using Brown-Forsythe and Welch ANOVA tests with Dunnett’s T3 multiple comparison test in order to correct for groups with significantly unequal variances. Error bars represented ± SD, unless stated otherwise. p < 0.05 was considered statistically significant.
Statistical analyses were performed with GraphPad Prism 9.0 (GraphPad Software, Inc., San Diego, CA).
CRediT authorship contribution statement
Harsh Maan: Conceptualization, Methodology, Data curation, Writing – review & editing. Tatyana L. Povolotsky: Conceptualization, Methodology, Data curation. Ziv Porat: . Maxim Itkin: Methodology. Sergey Malitsky: Methodology, Data curation. Ilana Kolodkin-Gal: Conceptualization, Methodology, Data curation, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.11.043.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hou Q, Kolodkin-Gal I. Harvesting the complex pathways of antibiotic production and resistance of soil bacilli for optimizing plant microbiome. FEMS Microbiol Ecol. 2020. [DOI] [PubMed]
- 2.Zhu H et al. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol. 2014;41 (2): 371-86. [DOI] [PubMed]
- 3.Arakawa K. Manipulation of metabolic pathways controlled by signaling molecules, inducers of antibiotic production, for genome mining in Streptomyces spp. Antonie Van Leeuwenhoek. 2018;111(5):743–751. doi: 10.1007/s10482-018-1052-6. [DOI] [PubMed] [Google Scholar]
- 4.Cornforth D.M., Foster K.R. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11(4):285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 5.Flemming H.-C., Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 2019;17(4):247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 6.Wilking J.N., et al. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2013;110(3):848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietrich L.E.P., et al. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195(7):1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom-Ackermann Z., et al. Toxin-Antitoxin systems eliminate defective cells and preserve symmetry in Bacillus subtilis biofilms. Environ Microbiol. 2016;18(12):5032–5047. doi: 10.1111/1462-2920.13471. [DOI] [PubMed] [Google Scholar]
- 9.Kolodkin-Gal I., et al. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 2013;27(8):887–899. doi: 10.1101/gad.215244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Toole G., et al. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54(1):49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 11.Sauer K., et al. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184(4):1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Southey-Pillig C.J., et al. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187(23):8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monds R.D., O’Toole G.A. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17(2):73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Lopez D., et al. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009;33(1):152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 15.Stewart P.S., Franklin M.J. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 16.Xu K.D., et al. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64(10):4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rani S.A., et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol. 2007;189(11):4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinberg N., Kolodkin-Gal I. The matrix reloaded: probing the extracellular matrix synchronizes bacterial communities. J Bacteriol. 2015 doi: 10.1128/JB.02516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branda S.S., et al. Biofilms: the matrix revisited. Trends Microbiol. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Keren-Paz A et al. A novel calcium-concentrating compartment drives biofilm formation and persistent infections. bioRxiv, 2020:2020.01.08.898569.
- 21.Keren-Paz A., Kolodkin-Gal I. A brick in the wall: Discovering a novel mineral component of the biofilm extracellular matrix. N Biotechnol. 2020;56:9–15. doi: 10.1016/j.nbt.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Arnaouteli S., et al. Bacillus subtilis biofilm formation and social interactions. Nat Rev Microbiol. 2021;19(9):600–614. doi: 10.1038/s41579-021-00540-9. [DOI] [PubMed] [Google Scholar]
- 23.Branda S.S., et al. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98(20):11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salinas N., et al. Emerging roles of functional bacterial amyloids in gene regulation, toxicity, and immunomodulation. Microbiol Mol Biol Rev. 2021;85(1) doi: 10.1128/MMBR.00062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romero D., et al. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107(5):2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branda S.S., et al. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59(4):1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi K., Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85(1):51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 28.Hobley L., et al. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A. 2013;110(33):13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cámara-Almirón J., et al. Dual functionality of the amyloid protein TasA in Bacillus physiology and fitness on the phylloplane. Nat Commun. 2020;11(1):1859. doi: 10.1038/s41467-020-15758-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinberg N., et al. The extracellular matrix protein TasA is a developmental cue that maintains a motile subpopulation within Bacillus subtilis biofilms. Sci Signal. 2020;13(632) doi: 10.1126/scisignal.aaw8905. [DOI] [PubMed] [Google Scholar]
- 31.Rubinstein S.M., et al. Osmotic pressure can regulate matrix gene expression in Bacillus subtilis. Mol Microbiol. 2012;86(2):426–436. doi: 10.1111/j.1365-2958.2012.08201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita M., et al. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187(4):1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vargas-Bautista C., et al. Bacterial competition reveals differential regulation of the pks genes by Bacillus subtilis. J Bacteriol. 2014;196(4):717–728. doi: 10.1128/JB.01022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian T., et al. Sucrose triggers a novel signaling cascade promoting Bacillus subtilis rhizosphere colonization. ISME J. 2021;15(9):2723–2737. doi: 10.1038/s41396-021-00966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beauregard P.B., et al. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A. 2013;110(17):E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuba-Surma E.K., et al. The ImageStream System: a key step to a new era in imaging. Folia Histochem Cytobiol. 2007;45(4):279–290. [PubMed] [Google Scholar]
- 37.Maan H., et al. Bacillus subtilis colonization of Arabidopsis thaliana roots induces multiple biosynthetic clusters for antibiotic production. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.722778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganin H., et al. Indole derivatives maintain the status quo between beneficial biofilms and their plant hosts. Mol Plant Microbe Interact. 2019;32(8):1013–1025. doi: 10.1094/MPMI-12-18-0327-R. [DOI] [PubMed] [Google Scholar]
- 39.Lopez D., et al. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A. 2009;106(1):280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López D., et al. Paracrine signaling in a bacterium. Genes Dev. 2009;23(14):1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maan H, et al. Resolving the conflict between antibiotic production and rapid growth by recognition of peptidoglycan of susceptible competitors. bioRxiv, 2021;2021.02.07.430110. [DOI] [PMC free article] [PubMed]
- 42.Carter M., Shieh J.C. Biochemical assays and intracellular signaling. Guide to Research Techniques in Neuroscience. 2010:297–329. [Google Scholar]
- 43.Magnuson R., et al. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77(2):207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 44.Kolter R., Greenberg E.P. Microbial sciences: the superficial life of microbes. Nature. 2006;441(7091):300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 45.Aguilar C., et al. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol. 2007;10(6):638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donlan R.M., Costerton J.W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costerton J.W., et al. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 48.Fux C.A., et al. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13(1):34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 49.Mah T.F., et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426(6964):306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 50.Stewart P.S. Biofilm accumulation model that predicts antibiotic resistance of Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 1994;38(5):1052–1058. doi: 10.1128/aac.38.5.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292(2):107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 52.Stewart P.S., Costerton J.W. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 53.Maunders E., Welch M. Matrix exopolysaccharides; the sticky side of biofilm formation. FEMS Microbiol Lett. 2017;364(13) doi: 10.1093/femsle/fnx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross P., et al. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55(1):35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ude S., et al. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol. 2006;8(11):1997–2011. doi: 10.1111/j.1462-2920.2006.01080.x. [DOI] [PubMed] [Google Scholar]
- 56.Serra D.O., et al. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol. 2013;195(24):5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonas K., et al. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 2007;7(1) doi: 10.1186/1471-2180-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Solano C, et al. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol 2002;43 (3): 793-808. [DOI] [PubMed]
- 59.Dragoš A., Kovács Á.T. The peculiar functions of the bacterial extracellular matrix. Trends Microbiol. 2017;25(4):257–266. doi: 10.1016/j.tim.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 60.Chai Y., et al. Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. MBio. 2012;3(4) doi: 10.1128/mBio.00184-12. e00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roux D., et al. Identification of Poly-N-acetylglucosamine as a major polysaccharide component of the Bacillus subtilis biofilm matrix. J Biol Chem. 2015;290(31):19261–19272. doi: 10.1074/jbc.M115.648709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kobayashi K. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J Bacteriol. 2007;189(13):4920–4931. doi: 10.1128/JB.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butcher R.A., et al. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104(5):1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Straight P.D., et al. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of surfactants in raising aerial structures. J Bacteriol. 2006;188(13):4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stubbendieck R.M., et al. Escape from lethal bacterial competition through coupled activation of antibiotic resistance and a mobilized subpopulation. PLoS Genet. 2015;11(12):e1005722. doi: 10.1371/journal.pgen.1005722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stubbendieck R.M., Straight P.D. Multifaceted interfaces of bacterial competition. J Bacteriol. 2016;198(16):2145–2155. doi: 10.1128/JB.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arima K., et al. Surfactin a crystalline peptidelipid surfactant produced by Bacillus subtilis - isolation characterization and its inhibition of fibrin clot formation. Biochem Biophys Res Commun. 1968;31(3):488–494. doi: 10.1016/0006-291x(68)90503-2. [DOI] [PubMed] [Google Scholar]
- 68.Rosenberg G., et al. Not so simple, not so subtle: the interspecies competition between Bacillus simplex and Bacillus subtilis and its impact on the evolution of biofilms. NPJ Biofilms Microbiomes. 2016;2(1):15027. doi: 10.1038/npjbiofilms.2015.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Danevcic T., et al. Surfactin facilitates horizontal gene transfer in Bacillus subtilis. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.657407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Povolotsky T.L., et al. Metabolic microenvironments drive microbial differentiation and antibiotic resistance. Trends Genet. 2021;37(1):4–8. doi: 10.1016/j.tig.2020.10.007. [DOI] [PubMed] [Google Scholar]
- 71.Adams J.C., Watt F.M. Regulation of development and differentiation by the extracellular matrix. Development. 1993;117(4):1183–1198. doi: 10.1242/dev.117.4.1183. [DOI] [PubMed] [Google Scholar]
- 72.Wilson G.A., Bott K.F. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1968;95(4):1439–1449. doi: 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farzand A., et al. Marker assisted detection and LC-MS analysis of antimicrobial compounds in different Bacillus strains and their antifungal effect on Sclerotinia sclerotiorum. Biol Control. 2019;133:91–102. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.