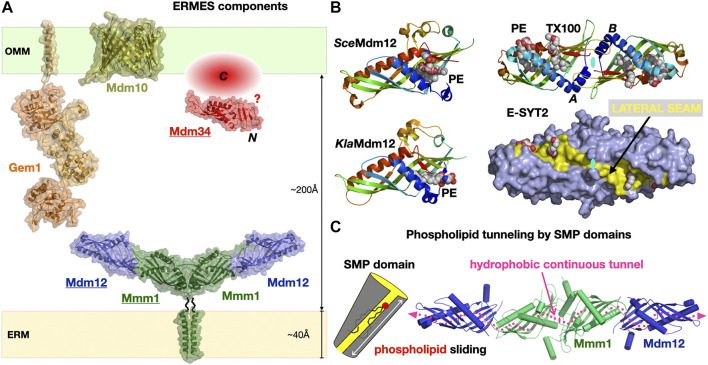
FIGURE 6.
The Endoplasmic Reticulum-to-Mitochondrion Encounter Structure. (A) Composite structural model of the yeast ERMES including the structures of the Mdm12/Mmm1 heterotetrameric core of SMP domains and the Mmd10 porin and combining the homology models of the Gem1 Miro GTPase and the Mdm34 SMP domain generated in Phyre 2 . The C-terminal domain of Mdm34 is represented as an unstructured module. (B) Structures of diverse SMP domains -Mdm12 (S. cerevisiae and K. lactis) and dimer of E-SYT2- showing the different positions sampled by bound PLs along the lateral seam of the SMP domain; the E-SYT2 dimer is represented using ribbon and also full surface (slate blue) representations to emphasize the existence of a lateral seam (yellow). PL ligands are shown as spheres; in E-SYT2 an additional molecule of Triton X100 detergent was observed. All structures are shown under the same orientation relative to E-SYT2 monomer A. (C) Hydrophobic tunnel spanning the entire length of the Mdm12/Mmm1 tetrameric assembly of aligned SMP domains for the sliding of PLs along the solvent-accessible lateral seam (magenta dotted line) exposing the polar headgroups of the PLs (PDB 5GYD-5H5A-4P42-5YK7).