Abstract
A massive depletion of CD4+ T lymphocytes has been described in early and acute human immunodeficiency virus (HIV) infection, leading to an imbalance between the human microbiome and immune responses. In recent years, a growing interest in the alterations in gut microbiota in HIV infection has led to many studies; however, only few studies have been conducted to explore the importance of oral microbiome in HIV-infected individuals. Evidence has indicated the dysbiosis of oral microbiota in people living with HIV (PLWH). Potential mechanisms might be related to the immunodeficiency in the oral cavity of HIV-infected individuals, including changes in secretory components such as reduced levels of enzymes and proteins in saliva and altered cellular components involved in the reduction and dysfunction of innate and adaptive immune cells. As a result, disrupted oral immunity in HIV-infected individuals leads to an imbalance between the oral microbiome and local immune responses, which may contribute to the development of HIV-related diseases and HIV-associated non-acquired immunodeficiency syndrome comorbidities. Although the introduction of antiretroviral therapy (ART) has led to a significant decrease in occurrence of the opportunistic oral infections in HIV-infected individuals, the dysbiosis in oral microbiome persists. Furthermore, several studies with the aim to investigate the ability of probiotics to regulate the dysbiosis of oral microbiota in HIV-infected individuals are ongoing. However, the effects of ART and probiotics on oral microbiome in HIV-infected individuals remain unclear. In this article, we review the composition of the oral microbiome in healthy and HIV-infected individuals and the possible effect of oral microbiome on HIV-associated oral diseases. We also discuss how ART and probiotics influence the oral microbiome in HIV infection. We believe that a deeper understanding of composition and function of the oral microbiome is critical for the development of effective preventive and therapeutic strategies for HIV infection.
Keywords: HIV, Oral microbiome, Antiretroviral therapy, Probiotics, Intervention
Introduction
Human immunodeficiency virus (HIV) infection is characterized by severe deficiency of the host immune system through the massive depletion of CD4+ T cells. The World Health Organization reported that, in the end of 2019, approximately, 38 million people were living with HIV worldwide, with around 67% of them receiving antiretroviral therapy (ART). Despite effective ART, several oral diseases, such as oropharyngeal candidiasis (OPC)[1,2] and periodontitis,[3,4] are frequently reported in all stages of HIV infection. In addition, as the life expectancy in people living with HIV (PLWH) increases, the risk of HIV-associated non-acquired immune deficiency syndrome (AIDS) comorbidities such as cardiovascular disease, neurocognitive disorders, cancer, and liver and kidney disease is increasingly reported.[5–7] Ryder et al[8] also found that older HIV-infected individuals who have received ART may present with a higher incidence of age-related oral diseases.
In recent years, several studies have shown that the composition of the gut microbiome in PLWH differs from that of HIV-uninfected individuals, including an increase in the abundance of Prevotella and a decrease in the abundance of Bacteroides.[9–12] Alterations in the gut microbiome may promote HIV-associated inflammation and immune activation.[13,14] Similarly, Annavajhala et al[15] suggested that oral microbiome diversity may also play a critical role in systemic inflammation in HIV-infected individuals. Studies have found that CD4+ T lymphocytes in gut-associated lymphoid tissue are greatly reduced in the early stage of HIV infection,[16] resulting in the loss of T helper (Th) 17 cell subsets.[17] It is believed that these interleukin-17- and interleukin-22-producing cells are essential to maintain intestinal epithelial integrity and gastrointestinal barrier function. Therefore, the loss of Th17 cells may contribute to microbial translocation from the gut mucosa into the systemic circulation, promoting inflammation and immune activation in HIV-infected adults.[18–20] Studies have indicated that Th17 cells are essential for the control of fungal colonization in the oral mucosa.[21–24] The structure and network of the oral mucosal immune system have also been described to be similar to those of the gastrointestinal mucosal immune system.[25] This accumulating evidence suggests that oral microbiota may be similar to gut microbiota, both of which might induce systemic diseases through systemic translocation in HIV infection. In addition, Schmidt et al[26] found that oral species, including opportunistic pathogens, might diffuse from the oral cavity to the gut, which may directly cause inflammation in the gut. Therefore, a focused effort on the effects of the oral microbiome on HIV will be critical.
Culture-dependent methods such as growth on media, microscopic observation, and biochemical analysis have been used to determine the composition of the microbiome. However, the appropriate culture conditions of some species might remain unclear, making them difficult to be cultivated. Currently, with the development of molecular techniques, next-generation sequencing, such as whole-metagenome shotgun sequencing and 16S ribosomal RNA amplicon sequencing, has been widely used for microbiome analysis. These “novel” technical approaches clearly contribute to the monitoring and manipulation of the human microbiome and provide new opportunities for diagnostics and therapeutics of human diseases.[27] To comprehensively understand the human microbiome and the relationship between the microbiome and human diseases, the National Institutes of Health of the US launched the “Human Microbiome Project” in 2007.[28] This review discusses alterations in the oral microbiome in HIV infection and the effects of the oral microbiome on HIV-associated oral diseases and evaluates the effects of potential interventions on the oral microbiome in HIV-infected individuals.
The human oral microbiome
The oral microbiome is an important part of the human microbiome and includes different microbes e.g., bacteria, fungi, viruses, mycoplasma, and protozoa.[29] Bacteria are the predominant group of the oral microbiome, of which approximately 700 bacterial species have been identified. The oral microbiome plays a critical role in human metabolism, physiology, and immunity, including inhibiting pathogenic microorganism colonization, maintaining the acid-base balance, regulating local oral immunity, and participating in salivary nitrate metabolism.[30]
The Human Oral Microbiome Database (http://www.homd.org/) contains records for a total of 775 microbial species. Among them, approximately 57% have been cultivated and named, 13% can be cultivated but not named, and 30% are uncultivated. Many of them have unique living conditions, such as specific temperature, pH, nutrition, and interaction with other species. The inability to fully replicate the ecological conditions in the oral cavity may be the reason why some oral microbiota cannot be cultivated artificially. The human oral bacterial microbiome consists primarily of six phyla: Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirochaetes, and Fusobacteria.[29,31] Bik et al[32] analyzed the bacterial diversity in the oral cavity of ten healthy individuals and found that the most abundant genus was Streptococcus, followed by Haemophilus, Neisseria, Prevotella, Veillonella, and Rothia, similar to other reports on the oral microbiome.[31,33,34]
In addition to bacterial communities, different fungi are also widely colonized in the human oral cavity.[35] It is known that in elderly and immunocompromised individuals, oral commensal fungi can also serve as opportunistic pathogens. Ghannoum et al[36] used internal transcribed spacer (ITS) sequencing to characterize the oral mycobiome in healthy individuals. This study demonstrated that the oral mycobiome comprises 74 culturable and 11 nonculturable fungal genera. In the samples from 20 healthy individuals, the most common genera were Candida (75%), followed by Cladosporium (65%), Aureobasidium and Saccharomycetales (50% for both), Aspergillus (35%), Fusarium (30%), and Cryptococcus (20%).
The alterations in the oral microbiome in HIV infection
The homeostasis of oral microbiota can be affected by multiple factors, including diet, smoking, and drugs. Moreover, changes in secretory components in saliva, innate and adaptive immune responses, and the physiological structure and function of the oral mucosa can also cause the dysbiosis of the oral microbiome.[37] Indeed, several studies have demonstrated a significant difference in the alterations of the oral microbiome between PLWH and HIV-uninfected healthy controls.[38–42] However, the potential mechanisms of oral microbiota changes in HIV infection remain unclear.
Alterations in salivary composition and function in HIV infection might play a key role in the dysbiosis of the oral microbiome. Saliva contains a variety of secretory components that are essential for maintaining oral homeostasis.[43] Studies have shown that secretory components, including immunoglobulin A (IgA), lysozyme, and host defense peptides, such as antimicrobial peptides, defensins, and histones, play an important role in microbial control and oral mucosal immunity.[44,45]
A recent study indicated the composition and function of saliva change in HIV infection. The impairment of local immunity in HIV infection, including decreased salivary IgA, defensins, and cytokines, might convert commensal microorganisms to microorganisms with increased pathogenicity and lead to the dysbiosis of oral microbiota, which could increase the risk of opportunistic infections.[46,47] Arirachakaran et al[48] showed that HIV-infected individuals, regardless of whether they are receiving ART, have a higher frequency and load of opportunistic microorganisms than HIV-uninfected controls. Other studies also revealed that the diversity and bacterial load in salivary samples from HIV-infected individuals were significantly higher than those in HIV-uninfected samples.[49] In addition, a negative correlation between oral lesions and CD4+ T-cell counts has been reported.[50–52] Therefore, the disruption of oral mucosal immunity in HIV infection might destroy the colonization of commensal bacteria in the oral cavity and lead to an increase in oral microbial diversity, resulting in an increased risk of HIV-associated oral diseases. However, different studies have also shown differing results. They found that the oral bacterial diversity in PLWH was significantly decreased when compared with that in HIV-uninfected individuals.[38,39,46,53] A possible explanation might be the increased proportion of opportunistic pathogens caused by immunodeficiency in HIV infection.[46] Furthermore, a variety of salivary proteins, such as lysozyme, defensin, lactoferrin, secretory leukocyte protease inhibitor, and salivary agglutinin, have been confirmed to inhibit HIV infectivity in vitro, which also shows the importance of changes in salivary components in the pathogenesis of HIV infection.[54]
In addition to the secretory components, cellular innate immune components in the oral cavity, e.g., macrophages, natural killer cells, polymorphonuclear leukocytes, and dendritic cells, also have the capacity to protect the oral mucosa from the colonization of pathogenic microorganisms.[55] These innate immune cells can recognize pathogens such as bacteria, viruses, and fungi through pattern recognition receptors (PRRs). PRRs mainly comprise several families, including Toll-like receptors, C-type lectin receptors, retinoic acid inducible gene-like receptors, and nucleotide-binding oligomerization domain-like receptors.[56] The binding of these PRRs to the pathogen-associated molecular patterns presented on the surface of microorganisms will induce the production of cytokines, chemokines, and vasoactive molecules, which might play important roles in regulating the innate immune response to infection and promoting the induction of adaptive immune responses.[55,57] Therefore, these innate immune cells are essential for the prevention of bacterial infections. However, innate immune responses in the oral cavity are impaired in HIV infection, which may lead to the dysbiosis of the oral microbiome and increase the occurrence of opportunistic infections.[44]
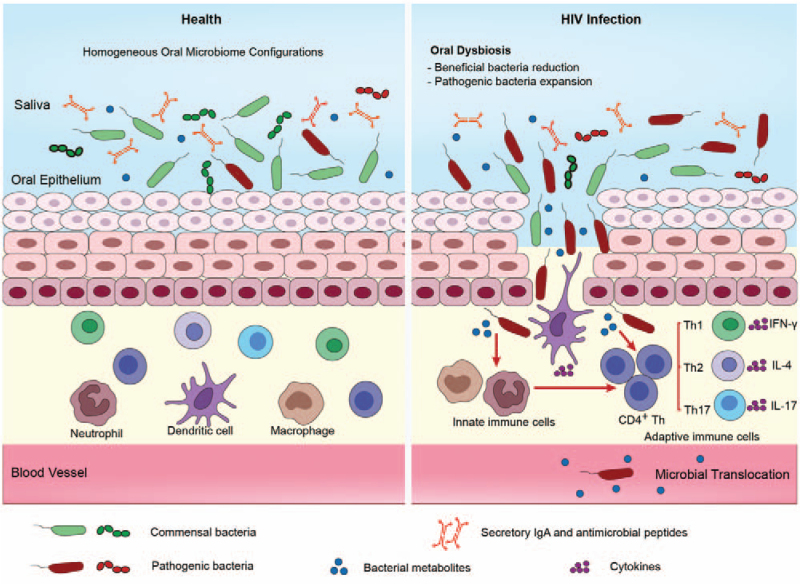
In addition, immunodeficiency associated with HIV infection might lead to defects in the adaptive immune response, which could also promote oral microbiota dysbiosis. A number of studies have shown that the Th17 immune response is essential for the control of fungal infection and inflammation of the oral mucosa.[21,58] Dutzan et al[59] found that Th17 cells could maintain oral barrier integrity and aid in fighting oral fungal infections. Furthermore, the critical role of Th17 cells in the fight against Candida infection has been described in a previous study.[60] In addition, Th1 cells might mediate early gingival inflammatory lesions in response to bacterial plaques by producing cytokines such as interferon γ (IFN-γ).[55,61] Th2 immune responses are also closely related to the progression of periodontal diseases[62] [Figure 1].
Figure 1.
Proposed mechanisms of oral microbiome dysbiosis in HIV infection. In health, oral epithelial cells have the capability to maintain microbial colonization. However, disrupted oral immunity, including changes in secretory components in saliva (sIgA, lysozyme, and antimicrobial peptides), deficiency of innate immune responses (macrophages and dendritic cells), and adaptive immune response (CD4+ Th), may cause oral microbiome dysbiosis in HIV infection. Such an imbalance between the oral microbiome and oral immune responses may also contribute to the development of HIV-related oral diseases (periodontal disease) and HIV-associated non-AIDS comorbidities. Periodontal disease is caused by the interplay between pathogenic bacteria and host defense, which can also lead to microbial translocation and an increased risk of systemic conditions. AIDS: Acquired Immune Deficiency Syndrome; HIV: Human immunodeficiency virus; IgA: Immunoglobulin A; IFN-γ: Interferon γ; IL-4: Interleukin-4; Th: T helper.
Although previous studies have demonstrated that oral microbiome composition might change in HIV infection, the results obtained from the different studies are not the same [Table 1]. A recent study compared saliva microbiome samples from HIV-infected individuals and HIV-uninfected controls. They found that the abundance of Streptococcus was increased in HIV-infected individuals, while the abundance of Neisseria was higher in healthy controls.[38] Another study showed similar results: the abundance of Veillonella, Rothia, and Streptococcus was significantly increased in the oral microbiome of PLWH, whereas the abundance of Neisseria was significantly decreased.[39] The oral microbiome in HIV-infected children and teenagers is also characterized by a higher frequency of the phyla Firmicutes and the genus Streptococcus.[63] However, other studies have shown that there are no large differences in the oral microbiome between HIV-treated patients and healthy controls.[64,65] In addition, alterations in the oral fungal community composition in PLWH have also been noted when compared with those in HIV-uninfected individuals.[66] A study compared the oral fungal composition of 12 HIV-infected individuals with that of 12 HIV-uninfected individuals. The study showed that Candida, Epicoccum, and Alternaria were the most common, presenting in 92%, 33%, and 25% of HIV-infected individuals, respectively. However, the most abundant fungi in HIV-uninfected controls were Candida, Pichia, and Fusarium, presenting in 58%, 33%, and 33%, respectively.[67] Fidel et al[68] have also demonstrated the relative abundance of oral fungal communities in 149 HIV-positive and 88 HIV-negative subjects. This study suggested that 168 species can be identified in 12 dominant genera by sequencing of the ITS2 region of the rRNA gene repeat. However, they indicated that the diversity of the oral mycobiome is usually dominated by a small number of species, and HIV and ART might affect the oral mycobiome composition. In addition, many studies have shown increased Candida colonization in HIV infection.[1,35,69,70] These studies support that HIV infection can significantly change the host oral microbiota. However, the effects of an altered oral microbiome on HIV-associated diseases remain to be shown.
Table 1.
The oral microbiome in HIV infection.
| Study | Cohort | Samples | Design | Major findings |
| Li et al[46] | 10 HIV+ subjects prior to and after 6 months of ART 10 HIV− controls | Saliva | Cross-sectional and longitudinal | • Increased oral streptococci, lactobacilli, Streptococcus mutans, and Candida in HIV • Increased Fusobacterium, Campylobacter, Prevotella, Capnocytophaga, Selenomonas, Actinomyces, Granulicatella, and Atopobium and decreased Aggregatibacter after ART |
| Beck et al[99] | 18 HIV+ naive 38 HIV+ ART 80 HIV− | Oral wash | Cross-sectional | • Enrichment of a Streptococcus Operational taxonomic unit (OTU) and two Actinomyces OTUs in HIV+ naive subjects, an Atopobium OTU in HIV+ naive and ART subjects, and a Rothia OTU in ART |
| Saxena et al[47] | 46 HIV+ subjects before and after ART and 69 HIV− controls 8 HIV+ subjects and 8 HIV− subjects | Saliva | Cross-sectional and longitudinal | • Different microbial compositions among HIV+ subjects before and after ART and HIV− controls • Differences in Streptococcacea, Prevotellaceae, Porphyromonadaceae, and Neisseriaceae between HIV+ and HIV− subjects |
| Arirachakaran et al[48] | 148 HIV+ ART for >5 years 20 HIV+ untreated 53 vertically transmitted ART HIV+ subjects 30 HIV− controls | Tongue, gingival crevices, and mucosal lesions | Cross-sectional | • Higher frequency and load of opportunistic microorganisms in the ART group and non-ART group than the HIV− controls • Increased Candida spp. on the tongue in HIV+ subjects with CD4+ counts <500 cells/mm3 |
| Presti et al[41] | 35 HIV+ subjects prior to and after 6 months of ART | Saliva | Longitudinal | • Higher bacterial richness and diversity in HIV+ subjects with persistently low CD4 counts after 24 weeks of ART • Differences in several taxa, including Porphyromonas species discriminated between HIV+ subjects before and after 6 months of ART |
| Starr et al[65] | 154 perinatally HIV-infected youth 100 perinatally HIV-exposed, uninfected youth | Subgingival plaque | Cross-sectional | • Similar microbiomes in the two cohorts • Two Corynebacterium species were lower in perinatally HIV-infected youth |
| Mukherjee et al[53] | 48 HIV-infected smokers 24 HIV-infected non-smokers 24 HIV-uninfected smokers | Oral wash | Cross-sectional | • Decreased Firmicutes and increased Proteobacteria in HIV-infected non-smokers compared to HIV-infected smokers and HIV-uninfected smokers • No difference in fungal phyla between the three cohorts |
| Gonçalves et al[63] | 27 HIV+ Brazilian children/teenagers 30 HIV− children/ teenagers | Whole saliva, biofilm from the dorsal surface of the tongue, and biofilm from supragingival and subgingival sites | Cross-sectional | • Higher frequency of the phyla Firmicutes and genus Streptococcus in HIV-infected children/teenagers • More complexity in oral microbiome of HIV-infected children/teenagers |
| Jiménez-Hernández et al[49] | 12 viremic ART-untreated 18 immunological ART responders 9 immunological ART non-responders 14 HIV-uninfected controls | Saliva | Cross-sectional and longitudinal | • Higher abundance of potential pathogens as Streptococcus agalactiae, Corynebacterium durum, and OTUs assigned to species of Prevotella, Leptotrichia, Tannerella, and Catonella in HIV+ subjects • Increased Actinobacteria, Rothia mucilaginosa, Mogibacterium and decreased Corynebacterium, Fusobacterium, and Prevotella melaninogenica in viremic ART-untreated subjects after prebiotics |
| Coker et al[95] | 94 HIV+ children 98 HIV exposed-uninfected children 94 HIV unexposed/ uninfected children | Saliva | Cross-sectional | • Depletion of eight bacterial taxa, including Actinomyces and Neisseria subflava and enrichment of Corynebacterium diphtheriae in HIV+ children when compared to HIV unexposed/uninfected children • Low CD4 levels persistently alter the oral microbiota. |
| Griffen et al[40] | 252 HIV+ ART 89 HIV− | Oral rinse | Cross-sectional | • A complex set of clinical features that influenced oral bacterial community composition, including the presence of HIV under ART |
| Yang et al[39] | 75 HIV+ ART 93 HIV− | Saliva | Cross-sectional | • Increased Veillonella, Rothia, and Streptococcus and decreased Neisseria |
| Annavajhala et al[15] | 52 HIV+ ART | Saliva and plaque samples | Cross-sectional and longitudinal | • Bacterial and fungal oral microbiome communities were associated with chronic systemic immune activation in HIV. |
| Li et al[38] | 20 HIV+ subjects before and after 6 months ART 20 HIV− controls | Saliva | Cross-sectional and longitudinal | • Increased Streptococcus and decreased Neisseria in HIV+ subjects • Provotella_7, Neisseria, and Haemophilus negatively correlated with CD4+ T cell count, while Neisseria positively correlated with viral load. |
| Imahashi et al[64] | 20 HIV+ Japanese with ART 13 HIV− controls | Saliva | Cross-sectional and longitudinal | • No largely differences in three major genera, Prevotella, Streptococcus, and Veillonella between the HIV+ subjects and controls. |
| Fidel et al[68] | 149 HIV+ subjects 88 HIV− subjects | Oral rinse | Cross-sectional | • Limited number of species dominated oral mycobiome • Several clinical variables, including HIV positivity and highly active antiretroviral therapy (HAART) affected the oral mycobiome |
| Li et al[42] | 15 acute HIV-infected subjects before and after ART 15 chronic HIV-infected subjects before and after ART 15 HIV− controls | Throat swabs | Cross-sectional and longitudinal | • Increased Prevotella in acute HIV infections and Streptococcus in chronic HIV infections • After effective ART, enriched Bradyrhizobium in both acute and chronic HIV infections, enriched Lactobacillus, Rothia, Clostridia, Actinobacteria, and Ruminococcaceae in controls. |
ART: Antiretroviral therapy; HIV: human immunodeficiency virus.
Effects of the oral microbiome on HIV-associated oral diseases
The oral microbiome is known to play an important role in host health and disease. On the one hand, the dysbiosis of the oral microbiome has been found in people with various oral diseases, such as dental caries, periodontal diseases, oral mucosal diseases, and oral cancer.[71] On the other hand, the dysbiosis of oral microbiota has also been observed in digestive system diseases (inflammatory bowel disease,[72–74] liver cirrhosis,[75] pancreatic cancer[76,77]), nervous system diseases (Alzheimer disease[78,79]), endocrine system diseases (diabetes[80]), immune system diseases (rheumatoid arthritis,[81,82] HIV infection[39,65]), cardiovascular diseases (arteriosclerosis[83]), adverse pregnancy outcomes,[84] and polycystic ovary syndrome.[85] The potential mechanisms may be that oral microbiota could enter the gastrointestinal tract and respiratory tract through eating and aspiration. Moreover, Han et al[86] reported that oral microbial dysbiosis could promote adverse systemic conditions through bacteremia. Studies have also shown that periodontal disease caused by the interaction between pathogenic microorganisms and host defenses can lead to microbial translocation and an increased risk of inflammatory diseases, such as cardiovascular diseases.[87,88] Therefore, the systemic translocation of the oral microbiome might also contribute to systemic diseases.
The oral cavity is one of the most common sites for opportunistic infections in PLWH. Several oral diseases often occur in PLWH, including periodontal diseases, OPC, oral warts, oral hairy leukoplakia, and Kaposi sarcoma (KS), even in those receiving ART.[44] Globally and throughout the decades, OPC has remained the most common oral manifestation in HIV infection, including among HIV-infected individuals receiving ART (26.2%).[89] OPC is caused by various Candida species, and Candida albicans is the most prevalent species isolated from PLWH.[70,90,91] The incidence of OPC in HIV infection is influenced by a multitude of factors, including immune status,[92] bacteriome-mycobiome interaction,[93] antifungal therapy, and ART.[94] Patil et al[1] showed that oral Candida colonization in HIV-infected patients increased, and it was significantly related to a lower CD4+ T-cell count. Coker et al[95] also demonstrated that low CD4+ T-cell levels in HIV-infected children might persistently alter oral microbiota. In addition, other studies found that the prevalence of dental caries and periodontal diseases in PLWH was increased, which may be related to changes in oral microbiota in HIV-infected patients.[3,96] Compared with PLWH with mild periodontal disease, Abiotrophia, Rothia, and unclassified Pasteurellaceae were enriched in HIV-infected individuals with moderate and severe periodontal disease, and Treponema spp. were enriched in patients with severe periodontal disease.[4] Moreover, Marion et al. found that oral microbiota composition in HIV-infected individuals with oral KS was significantly different from that of HIV-infected individuals without oral KS. They found that at the genus level, the abundances of Aggregatibacter and Lautropia were decreased, whereas those of Corynebacterium and Shuttleworthia were increased in HIV-infected individuals with oral KS.[97] These studies indicated that distinct oral microbiota might affect the development of oral diseases in PLWH.
Effects of potential interventions on the oral microbiome
Although the use of ART can inhibit HIV replication, increase CD4+ T lymphocytes, and reduce the occurrence of oral lesions, it cannot completely restore the oral microbiome of PLWH from its dysbiosis to normal.[38,98] Studies observed that the oral microbiome compositions in HIV-infected individuals with ART became more similar to those in HIV-uninfected controls; however, a difference remained between the groups.[38,99] Annavajhala et al[15] indicated that the use of specific ART regimens was associated with alterations in both gut and oral bacterial diversity. A study showed that Fusobacterium, Campylobacter, Prevotella, Capnocytophaga, Selenomonas, Actinomyces, Granulicatella, and Atopobium were increased in HIV-infected individuals after receiving ART, while Aggregatibacter was significantly decreased.[45] Another study collected samples from 35 HIV-infected subjects at baseline and after 24 weeks of ART to compare the differences in oral microbiota. The results showed that the dominant phyla in samples from patients with 24 weeks of ART remained similar to those observed at baseline, and the diversity was not significantly different between samples collected at baseline and those collected after 24 weeks of ART. However, PLWH with persistently low CD4+ T-cell counts had significantly increased bacterial richness and Shannon diversity, indicating that shifts in oral microbiota may play an important role in the recovery of CD4+ T-cell counts.[40] A recent study also found that Prevotella_7, Neisseria, and Haemophilus were negatively correlated with CD4+ T-cell count, whereas Neisseria was positively correlated with viral load.[37] Imahashi et al[64] also revealed the effects of long-term ART on gut and oral microbiota in PLWH. They suggested that ART, especially nucleoside reverse transcriptase inhibitor-based ART, has more suppressive effects on the composition and diversity of microbiota in the gut than that in the oral cavity.
Although OPC is still the most common oral opportunistic infection in PLWH, the introduction of ART can reduce its incidence in PLWH.[1] Interestingly, Maurya et al[100] found that although ART can decrease the risk of OPC in HIV-infected individuals, it does not decrease the colonization of Candida in the oral cavity. ART may play a key role in maintaining homeostasis between host immunity and the oral microbiome. However, it can also reduce oral commensal microorganisms that are capable of inhibiting pathogen colonization. Therefore, the potential mechanisms of ART on oral microbiome colonization remain unclear, and the further studies are necessary.
In addition to ART, many studies have demonstrated that probiotics can be used as a new therapeutic approach to improve the quality of life in PLWH.[101] Probiotics play a beneficial role in human health by regulating the immune system and controlling pathogen colonization.[102] Studies have shown that probiotics are effective in preventing and treating many disorders, such as acute gastroenteritis,[103] inflammatory bowel diseases and irritable bowel syndrome,[104,105]Clostridium difficile-associated diarrhea,[106] allergies,[107] neonatal sepsis,[108] and respiratory tract infections.[109] Hu et al[110] also reported that probiotics might exert their beneficial effects on coronavirus and have a positive effect on host immune functions during severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. In recent years, probiotics have also been used to prevent various oral diseases, such as dental caries,[111] gingivitis,[112] and periodontitis.[113] In addition, the administration of probiotics could have a beneficial effect on OPC.[114,115] De Barros et al[116] found that Lactobacillus could reduce the filamentation of C. albicans in in vitro and in vivo models, presenting the suppressive effect of probiotics on fungal pathogens. Other studies also showed that intake of Lactobacillus rhamnosus by immunosuppressed mice might decrease the development of candidiasis.[117] An in vitro study also suggested that Lactobacillus acidophilus and Lactobacillus plantarum had antifungal effects against different oral Candida species isolated from HIV/AIDS patients.[118] In addition, recent studies also focused on the intervention of prebiotics and found nutritional stimulation of beneficial bacteria by prebiotics might play a crucial role in promoting oral health.[119,120] Jiménez-Hernández et al[49] conducted a study on the impact of prebiotic intervention on the saliva microbiome of PLWH. A total of 32 HIV-infected subjects completed a 6-week prebiotic intervention, including viremic ART-untreated patients, immunological ART responders, immunological ART nonresponders, and HIV-uninfected controls. The diversity and richness of the saliva microbiome were decreased in the four groups after prebiotic intervention. In viremic ART-untreated individuals, the Actinobacteria Rothia mucilaginosa was increased, whereas some potential pathogens, such as Corynebacterium, Fusobacterium, or Prevotella melaninogenica, were decreased after the use of prebiotics. These studies have further demonstrated that the probiotics and prebiotics may be beneficial to regulate oral microbiota dysbiosis in HIV infection and prevent and reduce the occurrence of HIV-related oral diseases. However, the oral microbiome could be influenced by multiple factors. It is necessary to identify specific bacteria that are beneficial to preventing and treating HIV-related diseases. Further clinical studies are also needed to determine the efficacy and safety of probiotics in different clinical conditions.
Conclusion
Increasing evidence suggests a significant link of oral microbiome changes to HIV infection. We summarized the recent findings on alterations in oral microbiota of HIV infection and the potential roles of the shifts in oral microbiota in HIV-associated oral diseases. Moreover, we reviewed the effects of ART and probiotics on oral microbiota in HIV-infected individuals. It is evident that the oral microbiome plays an essential role in the pathogenesis of HIV disease, and a better understanding of the oral microbiome might improve the oral health of HIV-infected patients. In addition, further investigations are needed to evaluate the impact of the potential interventions on oral microbiome in HIV infection, which is groundwork for the formidable task of developing novel approaches for the prevention and therapy of HIV/AIDS-associated diseases.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 82072271, 81772165, and 81974303), the National 13th Five-Year Grand Program on Key Infectious Disease Control (Nos. 2017ZX10202101-004-001, 2017ZX10202102-005-003), the NSFC-NIH Biomedical collaborative research program (No. 81761128001), and the Beijing Key Laboratory for HIV/AIDS Research (No. BZ0089).
Conflicts of interest
None.
Footnotes
How to cite this article: Li S, Su B, He QS, Wu H, Zhang T. Alterations in the oral microbiome in HIV infection: causes, effects and potential interventions. Chin Med J 2021;134:2788–2798. doi: 10.1097/CM9.0000000000001825
References
- 1.Patil S, Majumdar B, Sarode SC, Sarode GS, Awan KH. Oropharyngeal candidosis in HIV-infected patients-an update. Front Microbiol 2018; 9:980.doi: 10.3389/fmicb.2018.00980. eCollection 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson GR, 3rd, Patel PK, Kirkpatrick WR, Westbrook SD, Berg D, Erlandsen J, et al. Oropharyngeal candidiasis in the era of antiretroviral therapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2010; 109:488–495. doi: 10.1016/j.tripleo.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polvora TLS, Nobre AVV, Tirapelli C, Taba M, Jr, de Macedo LD, Santana RC, et al. Relationship between human immunodeficiency virus (HIV-1) infection and chronic periodontitis. Expert Rev Clin Immunol 2018; 14:315–327. doi: 10.1080/1744666X.2018.1459571. [DOI] [PubMed] [Google Scholar]
- 4.Noguera-Julian M, Guillen Y, Peterson J, Reznik D, Harris EV, Joseph SJ, et al. Oral microbiome in HIV-associated periodontitis. Medicine (Baltimore) 2017; 96:e5821.doi: 10.1097/MD.0000000000005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhoades N, Mendoza N, Jankeel A, Sureshchandra S, Alvarez AD, Doratt B, et al. Altered immunity and microbial dysbiosis in aged individuals with long-term controlled HIV infection. Front Immunol 2019; 10:463.doi: 10.3389/fimmu.2019.00463. eCollection 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zicari S, Sessa L, Cotugno N, Ruggiero A, Morrocchi E, Concato C, et al. Immune activation, inflammation, and non-AIDS co-morbidities in HIV-infected patients under long-term ART. Viruses 2019; 11:200.doi: 10.3390/v11030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai SN, Landay AL. HIV and aging: role of the microbiome. Curr Opin HIV AIDS 2018; 13:22–27. doi: 10.1097/COH.0000000000000433. [DOI] [PubMed] [Google Scholar]
- 8.Ryder MI, Shiboski C, Yao TJ, Moscicki AB. Current trends and new developments in HIV research and periodontal diseases. Periodontol 2000 2020; 82:65–77. doi: 10.1111/prd.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocafort M, Noguera-Julian M, Rivera J, Pastor L, Guillen Y, Langhorst J, et al. Evolution of the gut microbiome following acute HIV-1 infection. Microbiome 2019; 7:73.doi: 10.1186/s40168-019-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armstrong AJS, Shaffer M, Nusbacher NM, Griesmer C, Fiorillo S, Schneider JM, et al. An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 2018; 6:198.doi: 10.1186/s40168-018-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol 2016; 9:24–37. doi: 10.1038/mi.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koay WLA, Siems LV, Persaud D. The microbiome and HIV persistence: implications for viral remission and cure. Curr Opin HIV AIDS 2018; 13:61–68. doi: 10.1097/COH.0000000000000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annavajhala MK, Khan SD, Sullivan SB, Shah J, Pass L, Kister K, et al. Oral and gut microbial diversity and immune regulation in patients with HIV on antiretroviral therapy. mSphere 2020; 5:e00798–e00819. doi: 10.1128/mSphere.00798-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Su B, Zhang X, Liu Y, Wu H, Zhang T. Incomplete immune reconstitution in HIV/AIDS patients on antiretroviral therapy: challenges of immunological non-responders. J Leukoc Biol 2020; 107:597–612. doi: 10.1002/JLB.4MR1019-189R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006; 12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 18.Mudd JC, Brenchley JM. Gut mucosal barrier dysfunction, microbial dysbiosis, and their role in HIV-1 disease progression. J Infect Dis 2016; 214:S58–S66. doi: 10.1093/infdis/jiw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 2015; 29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 20.Schuetz A, Deleage C, Sereti I, Rerknimitr R, Phanuphak N, Phuang-Ngern Y, et al. Initiation of ART during early acute HIV infection preserves mucosal Th17 function and reverses HIV-related immune activation. PLoS Pathog 2014; 10:e1004543.doi: 10.1371/journal.ppat.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez-Santos N, Huppler AR, Peterson AC, Khader SA, McKenna KC, Gaffen SL. Th17 cells confer long-term adaptive immunity to oral mucosal Candida albicans infections. Mucosal Immunol 2013; 6:900–910. doi: 10.1038/mi.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchner FR, LeibundGut-Landmann S. Tissue-resident memory Th17 cells maintain stable fungal commensalism in the oral mucosa. Mucosal Immunol 2021; 14:455–467. doi: 10.1038/s41385-020-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conti HR, Peterson AC, Brane L, Huppler AR, Hernandez-Santos N, Whibley N, et al. Oral-resident natural Th17 cells and gammadelta T cells control opportunistic Candida albicans infections. J Exp Med 2014; 211:2075–2084. doi: 10.1084/jem.20130877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med 2009; 206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu RQ, Zhang DF, Tu E, Chen QM, Chen WJ. The mucosal immune system in the oral cavity-an orchestra of T cell diversity. Int J Oral Sci 2014; 6:125–132. doi: 10.1038/ijos.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, et al. Extensive transmission of microbes along the gastrointestinal tract. Elife 2019; 8:e42693.doi: 10.7554/eLife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis JR, Gabaldon T. The human oral microbiome in health and disease: from sequences to ecosystems. Microorganisms 2020; 8:308.doi: 10.3390/microorganisms8020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, et al. NIH HMP Working Group. The NIH human microbiome project. Genome Res 2009; 19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Wang X, Li H, Ni C, Du Z, Yan F. Human oral microbiota and its modulation for oral health. Biomed Pharmacother 2018; 99:883–893. doi: 10.1016/j.biopha.2018.01.146. [DOI] [PubMed] [Google Scholar]
- 30.Marsh PD. In sickness and in health-what does the oral microbiome mean to us? An Ecological Perspective. Adv Dent Res 2018; 29:60–65. doi: 10.1177/0022034517735295. [DOI] [PubMed] [Google Scholar]
- 31.Lazarevic V, Whiteson K, Hernandez D, Francois P, Schrenzel J. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 2010; 11:523.doi: 10.1186/1471-2164-11-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 2010; 4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Contreras M, Costello EK, Hidalgo G, Magris M, Knight R, Dominguez-Bello MG. The bacterial microbiota in the oral mucosa of rural Amerindians. Microbiology (Reading) 2010; 156:3282–3287. doi: 10.1099/mic.0.043174-0. [DOI] [PubMed] [Google Scholar]
- 34.Mason MR, Chambers S, Dabdoub SM, Thikkurissy S, Kumar PS. Characterizing oral microbial communities across dentition states and colonization niches. Microbiome 2018; 6:67.doi: 10.1186/s40168-018-0443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sodre CS, Rodrigues PMG, Vieira MS, da Silva AMP, Goncalves LS, Ribeiro MG, et al. Oral mycobiome identification in atopic dermatitis, leukemia, and HIV patients - a systematic review. J Oral Microbiol 2020; 12:1807179.doi: 10.1080/20002297.2020.1807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 2010; 6:e1000713.doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samaranayake L, Matsubara VH. Normal oral flora and the oral ecosystem. Dent Clin N Am 2017; 61:199–215. doi: 10.1016/j.cden.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Chang S, Guo H, Ji Y, Jiang H, Ruan L, et al. Altered salivary microbiome in the early stage of HIV infections among young Chinese men who have sex with men (MSM). Pathogens 2020; 9:960.doi: 10.3390/pathogens9110960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Dunlap DG, Qin S, Fitch A, Li K, Koch CD, et al. Alterations in oral microbiota in HIV are related to decreased pulmonary function. Am J Respir Crit Care Med 2020; 201:445–457. doi: 10.1164/rccm.201905-1016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffen AL, Thompson ZA, Beall CJ, Lilly EA, Granada C, Treas KD, et al. Significant effect of HIV/HAART on oral microbiota using multivariate analysis. Sci Rep 2019; 9:19946.doi: 10.1038/s41598-019-55703-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Presti RM, Handley SA, Droit L, Ghannoum M, Jacobson M, Shiboski CH, et al. Alterations in the oral microbiome in HIV-infected participants after antiretroviral therapy administration are influenced by immune status. AIDS 2018; 32:1279–1287. doi: 10.1097/QAD.0000000000001811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Zhu J, Su B, Wei H, Chen F, Liu H, et al. Alteration in oral microbiome among men who have sex with men with acute and chronic HIV infection on antiretroviral therapy. Front Cell Infect Microbiol 2021; 11:695515.doi: 10.3389/fcimb.2021.695515. eCollection 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belstrom D. The salivary microbiota in health and disease. J Oral Microbiol 2020; 12:1723975.doi:10.1080/20002297.2020.1723975. eCollection 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heron SE, Elahi S. HIV infection and compromised mucosal immunity: oral manifestations and systemic inflammation. Front Immunol 2017; 8:241.doi: 10.3389/fimmu.2017.00241. eCollection 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nizamuddin I, Koulen P, McArthur CP. Contribution of HIV infection, AIDS, and antiretroviral therapy to exocrine pathogenesis in salivary and lacrimal glands. Int J Mol Sci 2018; 19:2747.doi: 10.3390/ijms19092747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Saxena D, Chen Z, Liu G, Abrams WR, Phelan JA, et al. HIV infection and microbial diversity in saliva. J Clin Microbiol 2014; 52:1400–1411. doi: 10.1128/JCM.02954-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saxena D, Li Y, Devota A, Pushalkar S, Abrams W, Barber C, et al. Modulation of the orodigestive tract microbiome in HIV-infected patients. Oral Dis 2016; 22:73–78. doi: 10.1111/odi.12392. [DOI] [PubMed] [Google Scholar]
- 48.Arirachakaran P, Poovorawan Y, Dahlen G. Highly-active antiretroviral therapy and oral opportunistic microorganisms in HIV-positive individuals of Thailand. J Investig Clin Dent 2016; 7:158–167. doi: 10.1111/jicd.12142. [DOI] [PubMed] [Google Scholar]
- 49.Jimenez-Hernandez N, Serrano-Villar S, Domingo A, Pons X, Artacho A, Estrada V, et al. Modulation of saliva microbiota through prebiotic intervention in HIV-infected individuals. Nutrients 2019; 11:1346.doi: 10.3390/nu11061346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saini R. Oral lesions: a true clinical indicator in human immunodeficiency virus. J Nat Sci Biol Med 2011; 2:145–150. doi: 10.4103/0976-9668.92316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vohra P, Jamatia K, Subhada B, Tiwari RVC, Althaf MN, Jain C. Correlation of CD4 counts with oral and systemic manifestations in HIV patients. J Fam Med Prim Care 2019; 8:3247–3252. doi: 10.4103/jfmpc.jfmpc_767_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frimpong P, Amponsah EK, Abebrese J, Kim SM. Oral manifestations and their correlation to baseline CD4 count of HIV/AIDS patients in Ghana. J Korean Assoc Oral Maxillofac Surg 2017; 43:29–36. doi: 10.5125/jkaoms.2017.43.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee PK, Chandra J, Retuerto M, Tatsuoka C, Ghannoum MA, McComsey GA. Dysbiosis in the oral bacterial and fungal microbiome of HIV-infected subjects is associated with clinical and immunologic variables of HIV infection. PLoS One 2018; 13:e0200285.doi: 10.1371/journal.pone.0200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corstjens PLAM, Abrams WR, Malamud D. Saliva and viral infections. Periodontol 2000 2016; 70:93–110. doi: 10.1111/prd.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feller L, Altini M, Khammissa RA, Chandran R, Bouckaert M, Lemmer J. Oral mucosal immunity. Oral Surg Oral Med Oral Pathol Oral Radiol 2013; 116:576–583. doi: 10.1016/j.oooo.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 57.McKernan DP. Pattern recognition receptors as potential drug targets in inflammatory disorders. Adv Protein Chem Struct Biol 2020; 119:65–109. doi: 10.1016/bs.apcsb.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Gaffen SL, Moutsopoulos NM. Regulation of host-microbe interactions at oral mucosal barriers by type 17 immunity. Sci Immunol 2020; 5:eaau4594.doi: 10.1126/sciimmunol.aau4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dutzan N, Abusleme L, Bridgeman H, Greenwell-Wild T, Zangerle-Murray T, Fife ME, et al. On-going mechanical damage from mastication drives homeostatic Th17 cell responses at the oral barrier. Immunity 2017; 46:133–147. doi: 10.1016/j.immuni.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bacher P, Hohnstein T, Beerbaum E, Rocker M, Blango MG, Kaufmann S, et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell 2019; 176:1340–1355. e15. doi: 10.1016/j.cell.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 61.Speakman EA, Dambuza IM, Salazar F, Brown GD. T cell antifungal immunity and the role of C-type lectin receptors. Trends Immunol 2020; 41:61–76. doi: 10.1016/j.it.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gemmell E, Yamazaki K, Seymour GJ. The role of T cells in periodontal disease: homeostasis and autoimmunity. Periodontol 2000 2007; 43:14–40. doi: 10.1111/j.1600-0757.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 63.Goncalves LS, de Carvalho Ferreira D, Heng NCK, Vidal F, Santos HF, Zanicotti DG, et al. Oral bacteriome of HIV-1-infected children from Rio de Janeiro, Brazil: next-generation DNA sequencing analysis. J Clin Periodontol 2019; 46:1192–1204. doi: 10.1111/jcpe.13176. [DOI] [PubMed] [Google Scholar]
- 64.Imahashi M, Ode H, Kobayashi A, Nemoto M, Matsuda M, Hashiba C, et al. Impact of long-term antiretroviral therapy on gut and oral microbiotas in HIV-1-infected patients. Sci Rep 2021; 11:960.doi: 10.1038/s41598-020-80247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Starr JR, Huang Y, Lee KH, Murphy CM, Moscicki AB, Shiboski CH, et al. Oral microbiota in youth with perinatally acquired HIV infection. Microbiome 2018; 6:100.doi: 10.1186/s40168-018-0484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hager CL, Ghannoum MA. The mycobiome in HIV. Curr Opin HIV AIDS 2018; 13:69–72. doi: 10.1097/COH.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, et al. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog 2014; 10:e1003996.doi: 10.1371/journal.ppat.1003996. eCollection 2014 Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fidel PL, Jr, Thompson ZA, Lilly EA, Granada C, Treas K, Dubois KR, 3rd, et al. Effect of HIV/HAART and other clinical variables on the oral mycobiome using multivariate analyses. mBio 2021; 12:e00294–e00321. doi: 10.1128/mBio.00294-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Armstrong-James D, Meintjes G, Brown GD. A neglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol 2014; 22:120–127. doi: 10.1016/j.tim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 70.Pour AH, Salari S, Almani PGN. Oropharyngeal candidiasis in HIV/AIDS patients and non-HIV subjects in the Southeast of Iran. Curr Med Mycol 2018; 4:1–6. doi: 10.18502/cmm.4.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell 2018; 9:488–500. doi: 10.1007/s13238-018-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang HN, Zhou XD, Xu X, Wang Y. Oral microbiota and inflammatory bowel disease (in Chinese). West China journal of stomatology 2019; 37:443–449. doi: 10.7518/hxkq.2019.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res 2014; 21:15–25. doi: 10.1093/dnares/dst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qi Y, Zang SQ, Wei J, Yu HC, Yang Z, Wu HM, et al. High-throughput sequencing provides insights into oral microbiota dysbiosis in association with inflammatory bowel disease. Genomics 2021; 113:664–676. doi: 10.1016/j.ygeno.2020.09.063. [DOI] [PubMed] [Google Scholar]
- 75.Acharya C, Sahingur SE, Bajaj JS. Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI Insight 2017; 2:e94416.doi: 10.1172/jci.insight.94416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012; 61:582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaiser RA, Halimi A, Alkharaan H, Lu L, Davanian H, Healy K, et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 2019; 68:2186–2194. doi: 10.1136/gutjnl-2018-317458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu YF, Lee WF, Salamanca E, Yao WL, Su JN, Wang SY, et al. Oral microbiota changes in elderly patients, an indicator of Alzheimer's Disease. Int J Environ Res Public Health 2021; 18:4211.doi: 10.3390/ijerph18084211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sureda A, Daglia M, Castilla SA, Sanadgol N, Nabavi SF, Khan H, et al. Oral microbiota and Alzheimer's disease: do all roads lead to Rome? Pharmacol Res 2020; 151:104582.doi: 10.1016/j.phrs.2019.104582. [DOI] [PubMed] [Google Scholar]
- 80.Casarin RCV, Barbagallo A, Meulman T, Santos VR, Sallum EA, Nociti FH, et al. Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J Periodontal Res 2013; 48:30–36. doi: 10.1111/j.1600-0765.2012.01498.x. [DOI] [PubMed] [Google Scholar]
- 81.Tong Y, Zheng L, Qing P, Zhao H, Li Y, Su L, et al. Oral microbiota perturbations are linked to high risk for rheumatoid arthritis. Front Cell Infect Microbiol 2019; 9:475.doi: 10.3389/fcimb.2019.00475. eCollection 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015; 21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 83.Liu XR, Xu Q, Xiao J, Deng YM, Tang ZH, Tang YL, et al. Role of oral microbiota in atherosclerosis. Clin Chim Acta 2020; 506:191–195. doi: 10.1016/j.cca.2020.03.033. [DOI] [PubMed] [Google Scholar]
- 84.Madianos PN, Bobetsis YA, Offenbacher S. Adverse pregnancy outcomes (APOs) and periodontal disease: pathogenic mechanisms. J Periodontol 2013; 84:S170–S180. doi: 10.1902/jop.2013.1340015. [DOI] [PubMed] [Google Scholar]
- 85.Li N, Li Y, Qian C, Liu Q, Cao W, Ma M, et al. Dysbiosis of the saliva microbiome in patients with polycystic ovary syndrome. Front Cell Infect Microbiol 2021; 10:624504.doi: 10.3389/fcimb.2020.624504. eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han YW, Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res 2013; 92:485–491. doi: 10.1177/0022034513487559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leishman SJ, Do HL, Ford PJ. Cardiovascular disease and the role of oral bacteria. J Oral Microbiol 2010; 2.doi: 10.3402/jom.v2i0.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Humphrey LL, Fu R, Buckley DI, Freeman M, Helfand M. Periodontal disease and coronary heart disease incidence: a systematic review and meta-analysis. J Gen Intern Med 2008; 23:2079–2086. doi: 10.1007/s11606-008-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El Howati A, Tappuni A. Systematic review of the changing pattern of the oral manifestations of HIV. J Investig Clin Dent 2018; 9:e12351.doi: 10.1111/jicd.12351. [DOI] [PubMed] [Google Scholar]
- 90.Khedri S, Santos ALS, Roudbary M, Hadighi R, Falahati M, Farahyar S, et al. Iranian HIV/AIDS patients with oropharyngeal candidiasis: identification, prevalence and antifungal susceptibility of Candida species. Lett Appl Microbiol 2018; 67:392–399. doi: 10.1111/lam.13052. [DOI] [PubMed] [Google Scholar]
- 91.Kwamin F, Nartey NO, Codjoe FS, Newman MJ. Distribution of Candida species among HIV-positive patients with oropharyngeal candidiasis in Accra, Ghana. J Infect Dev Ctries 2013; 7:41–45. doi: 10.3855/jidc.2442. [DOI] [PubMed] [Google Scholar]
- 92.de Repentigny L, Lewandowski D, Jolicoeur P. Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin Microbiol Rev 2004; 17:729–759. doi: 10.1128/CMR.17.4.729-759.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oever JT, Netea MG. The bacteriome-mycobiome interaction and antifungal host defense. Eur J Immunol 2014; 44:3182–3191. doi: 10.1002/eji.201344405. [DOI] [PubMed] [Google Scholar]
- 94.Patton LL. Current strategies for prevention of oral manifestations of human immunodeficiency virus. Oral Surg Oral Med Oral Pathol Oral Radiol 2016; 121:29–38. doi: 10.1016/j.oooo.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 95.Coker MO, Mongodin EF, El-Kamary SS, Akhigbe P, Obuekwe O, Omoigberale A, et al. Immune status, and not HIV infection or exposure, drives the development of the oral microbiota. Sci Rep 2020; 10:10830.doi: 10.1038/s41598-020-67487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Souza Goncalves L, Ferreira SMS, Souza CO, Souto R, Colombo AP. Clinical and microbiological profiles of human immunodeficiency virus (HIV)-seropositive Brazilians undergoing highly active antiretroviral therapy and HIV-seronegative Brazilians with chronic periodontitis. J Periodontol 2007; 78:87–96. doi: 10.1902/jop.2007.060040. [DOI] [PubMed] [Google Scholar]
- 97.Gruffaz M, Zhang T, Marshall V, Goncalves P, Ramaswami R, Labo N, et al. Signatures of oral microbiome in HIV-infected individuals with oral Kaposi's sarcoma and cell-associated KSHV DNA. PLoS Pathog 2020; 16:e1008114.doi: 10.1371/journal.ppat.1008114. eCollection 2020 Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lewy T, Hong BY, Weiser B, Burger H, Tremain A, Weinstock G, et al. Oral microbiome in HIV-infected women: Shifts in the abundance of pathogenic and beneficial bacteria are associated with aging, HIV load, CD4 count, and antiretroviral therapy. AIDS Res Hum Retroviruses 2019; 35:276–286. doi: 10.1089/AID.2017.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beck JM, Schloss PD, Venkataraman A, Twigg H, 3rd, Jablonski KA, Bushman FD, et al. Multicenter comparison of lung and oral microbiomes of HIV-infected and HIV-uninfected individuals. Am J Respir Crit Care Med 2015; 192:1335–1344. doi: 10.1164/rccm.201501-0128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maurya V, Srivastava A, Mishra J, Gaind R, Marak RSK, Tripathi AK, et al. Oropharyngeal candidiasis and Candida colonization in HIV positive patients in northern India. J Infect Dev Ctries 2013; 7:608–613. doi: 10.3855/jidc.2801. [DOI] [PubMed] [Google Scholar]
- 101.D’Angelo C, Reale M, Costantini E. Microbiota and probiotics in health and HIV infection. Nutrients 2017; 9:615.doi: 10.3390/nu9060615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Borchers AT, Selmi C, Meyers FJ, Keen CL, Gershwin ME. Probiotics and immunity. J Gastroenterol 2009; 44:26–46. doi: 10.1007/s00535-008-2296-0. [DOI] [PubMed] [Google Scholar]
- 103.Schnadower D, Tarr PI, Casper TC, Gorelick MH, Dean JM, O’Connell KJ, et al. Lactobacillus rhamnosus GG versus placebo for acute gastroenteritis in children. N Engl J Med 2018; 379:2002–2014. doi: 10.1056/NEJMoa1802598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jakubczyk D, Leszczynska K, Gorska S. The effectiveness of probiotics in the treatment of inflammatory bowel disease (IBD)-a Critical Review. Nutrients 2020; 12:1973.doi: 10.3390/nu12071973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol 2020; 145:16–27. doi: 10.1016/j.jaci.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 106.Goldenberg JZ, Yap C, Lytvyn L, Lo CKF, Beardsley J, Mertz D, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev 2017; (5):CD006095.doi:10.1002/14651858.CD006095.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.West CE, Dzidic M, Prescott SL, Jenmalm MC. Bugging allergy; role of pre-, pro- and synbiotics in allergy prevention. Allergol Int 2017; 66:529–538. doi: 10.1016/j.alit.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 108.Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med 2019; 25:716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 109.Hao Q, Dong BR, Wu T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst Rev 2015; (2):CD006895.doi: 10.1002/14651858. [DOI] [PubMed] [Google Scholar]
- 110.Hu J, Zhang L, Lin W, Tang W, Chan FKL, Ng SC. Review article: probiotics, prebiotics and dietary approaches during COVID-19 pandemic. Trends Food Sci Technol 2021; 108:187–196. doi: 10.1016/j.jpgs.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bustamante M, Oomah BD, Mosi-Roa Y, Rubilar M, Burgos-Diaz C. Probiotics as an adjunct therapy for the treatment of halitosis, dental caries and periodontitis. Probiotics Antimicrob Proteins 2020; 12:325–334. doi: 10.1007/s12602-019-9521-4. [DOI] [PubMed] [Google Scholar]
- 112.Montero E, Iniesta M, Rodrigo M, Marin MJ, Figuero E, Herrera D, et al. Clinical and microbiological effects of the adjunctive use of probiotics in the treatment of gingivitis: a randomized controlled clinical trial. J Clin Periodontol 2017; 44:708–716. doi: 10.1111/jcpe.12752. [DOI] [PubMed] [Google Scholar]
- 113.Invernici MM, Salvador SL, Silva PHF, Soares MSM, Casarin R, Palioto DB, et al. Effects of bifidobacterium probiotic on the treatment of chronic periodontitis: a randomized clinical trial. J Clin Periodontol 2018; 45:1198–1210. doi: 10.1111/jcpe.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mundula T, Ricci F, Barbetta B, Baccini M, Amedei A. Effect of probiotics on oral candidiasis: a systematic review and meta-analysis. Nutrients 2019; 11:2449.doi: 10.3390/nu11102449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chanda W, Joseph TP, Wang W, Padhiar AA, Zhong M. The potential management of oral candidiasis using anti-biofilm therapies. Med Hypotheses 2017; 106:15–18. doi: 10.1016/j.mehy.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 116.de Barros PP, Scorzoni L, de Camargo Ribeiro F, de Oliveira Fugisaki LR, Fuchs BB, Mylonakis E, et al. Lactobacillus paracasei 28.4 reduces in vitro hyphae formation of Candida albicans and prevents the filamentation in an experimental model of Caenorhabditis elegans. Microb Pathog 2018; 117:80–87. doi: 10.1016/j.micpath.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 117.Leao MVP, Tavares TAA, Goncalves ESCR, Dos Santos SSF, Junqueira JC, de Oliveira LD, et al. Lactobacillus rhamnosus intake can prevent the development of Candidiasis. Clin Oral Investig 2018; 22:2511–2518. doi: 10.1007/s00784-018-2347-8. [DOI] [PubMed] [Google Scholar]
- 118.Salari S, Almani PGN. Antifungal effects of Lactobacillus acidophilus and Lactobacillus plantarum against different oral Candida species isolated from HIV/AIDS patients: an in vitro study. J Oral Microbiol 2020; 12:1769386.doi: 10.1080/20002297.2020.1769386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Slomka V, Hernandez-Sanabria E, Herrero ER, Zaidel L, Bernaerts K, Boon N, et al. Nutritional stimulation of commensal oral bacteria suppresses pathogens: the prebiotic concept. J Clin Periodontol 2017; 44:344–352. doi: 10.1111/jcpe.12700. [DOI] [PubMed] [Google Scholar]
- 120.Slomka V, Herrero ER, Boon N, Bernaerts K, Trivedi HM, Daep C, et al. Oral prebiotics and the influence of environmental conditions in vitro. J Periodontol 2018; 89:708–717. doi: 10.1002/JPER.17-0437. [DOI] [PubMed] [Google Scholar]