Figure 1. Fasting triggers a ketogenic program in ISC and TA cells.
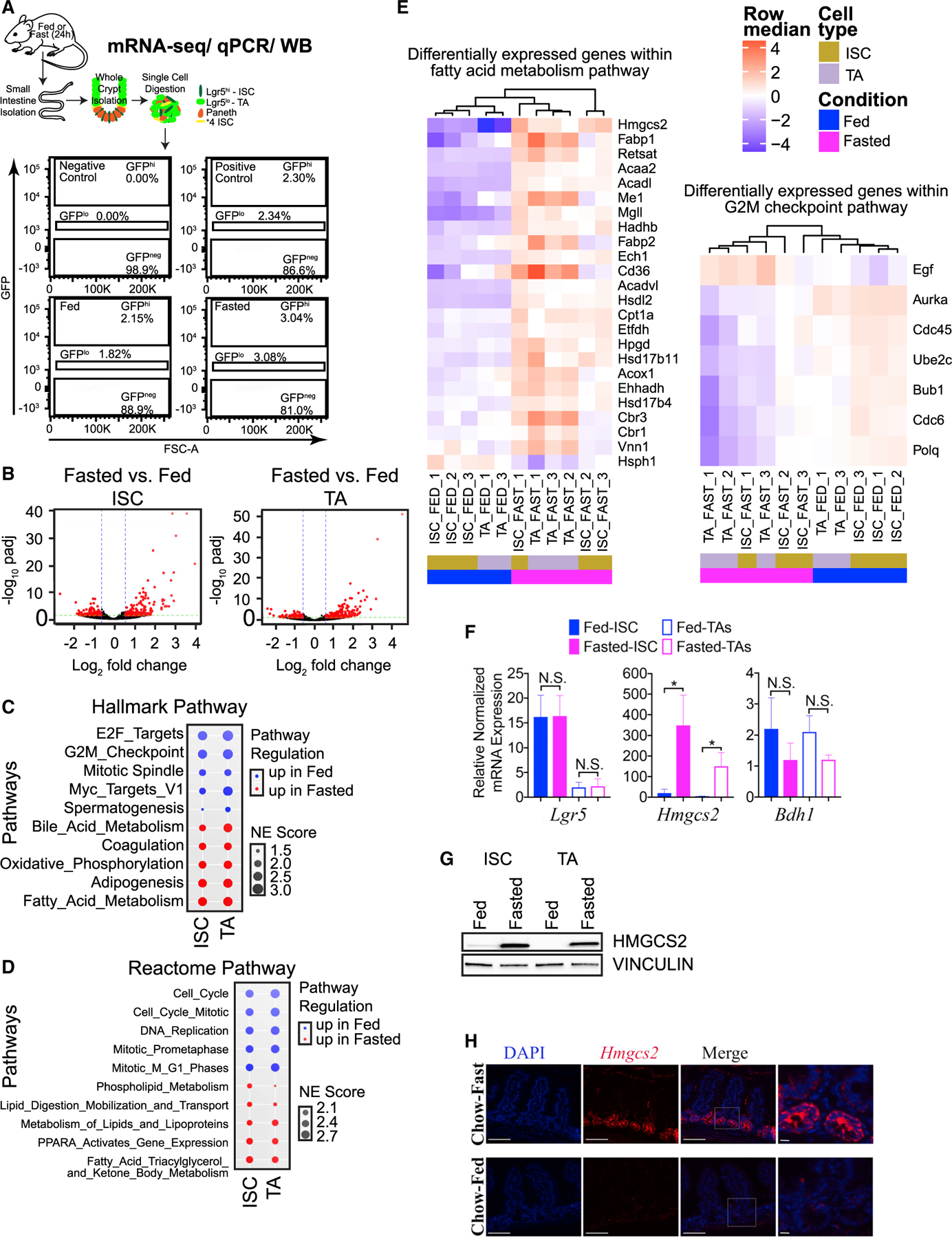
(A) Schematic of Lgr5+ cell collection (ISCs or TA cells) from SI crypts for ultra-low mRNA-seq (20,000 total cells per sample), qPCR (5,000 total cells per sample), or western blot (20,000 total cells per sample) analyses. Representative flow cytometry plots are included. FSC, forward scatter parameter.
(B) Volcano plots identify (304) differentially enriched genes by fasting (up- and downregulated) in ISCs (Lgr5−GFPhi) and (401) differentially enriched genes from matched TA progenitor cells (Lgr5−GFPlo). n = 3 mice per group.
(C) Top 5 up- or downregulated HALLMARK pathways enriched by fasting in ISCs (Lgr5−GFPhi) and TA cells (Lgr5−GFPlo), ranked by normalized enrichment (NE) score prevalence in ISCs.
(D) Top 5 up- or downregulated Reactome pathways enriched by fasting in ISCs (Lgr5−GFPhi) and TA cells (Lgr5−GFPlo), ranked by NE score prevalence in ISCs.
(E) Heatmap of RNA-seq analysis, showing relative expression of “fatty acid metabolism” and “G2M checkpoint” HALLMARK gene collections in ISCs and TA cells from fed and fasted replicate mice. All genes displayed are differentially expressed (false discovery rate [FDR] ≤ 0.05, log2FC > = 1). n = 3 mice per group.
(F) qPCR analysis confirming that Lgr5+ ISCs and TA cells were isolated by Lgr5 expression and fasting responsive gene expression of Hmgcs2 in ISCs and TA cells and Bdh1 in TA cells. Gapdh was multiplexed and used as a housekeeping gene per sample. n = 3 experiments, n = 3 mice per group. Data are mean ± SEM, analyzed by Student’s t test, unpaired. N.S., not significant.
(G) Representative western blot of HMGCS2 in ISCs and matched TAs of fed and fasted mice. VINCULIN was used as a loading control for 20,000 cells per sample. n = 4 experiments (n = 6–8 total mice per group).
(H) Immunofluorescence analysis of HMGCS2 in SI tissues (jejunum) of fed and fasted mice. Representative images from all groups are shown. Scale bars, 100 μm. Magnification, 40×. Magnified images, 40 μm.
See also Figure S1.
