Figure 3. Histone Kbhb is associated with an active chromatin landscape.
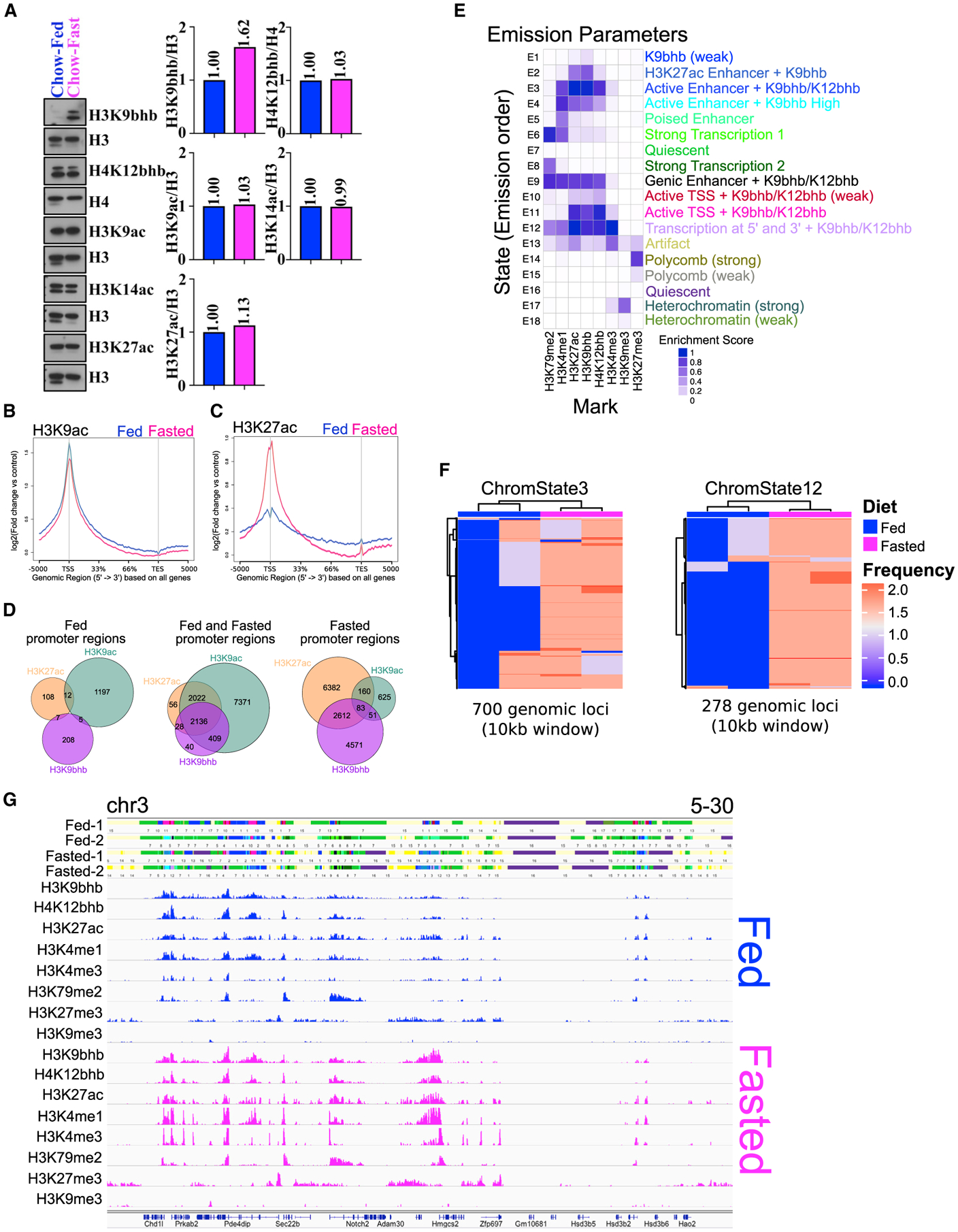
(A) Western blot analysis of histones isolated from small intestinal crypts probed with histone antibodies for H3K9bhb, H4K12bhb, H3K9ac, H3K14ac, and H3K27ac under fed and fasted conditions. Antibodies against H3 and H4 were used as loading controls. The relative quantification for all antibodies was normalized to total levels of H3 or H4.
(B and C) Signal intensity gene body plots of H3K9ac and H3K27ac based on all Ensembl genes under fed and fasted conditions. Ensembl genes are arranged 5′ to 3′ from −5 kb to the TSS, TSS, 33%, 66%, TES, and +5 kb from the TES. An arrow denotes the TSS at the 5′ region of the gene.
(D) Co-localization analysis of H3K9bhb + H3K9ac + H3K27ac consensus peaks within proximal promoter regions (±5 kb of the TSS) of SI crypt cells under the fed and fasted conditions.
(E) ChromHMM chromatin state profile of SI crypts from 1-kb interval genome bin size. User assigned functionalization of emission parameters is included.
(F) Unsupervised clustering analysis of the most variable regions for active chromatin state E3_active enhancers (marked by high levels of H3K27ac and H3K4me1 outside of the TSS) and E12_transcription at 5′ and 3′ (marked by high levels of H3K4me3, H3K27ac, H3K4me1, and H3K79me2) under fed and fasted conditions (two biological replicates in each). Frequency is the occurrence of the specified chromatin state within a 10-kb genomic window.
(G) 18-state ChromHMM model annotations in individual fed and fasted replicates around the Hmgcs2 locus, with each emission state marked by a separate color within the track. For each 1-kb interval, the corresponding input consensus ChIP-seq peak profiles of each histone mark utilized for ChromHMM binary response calls are shown for fed (blue) and fasted (magenta) sequence tag mark profiles.
See also Figures S3–S5.
