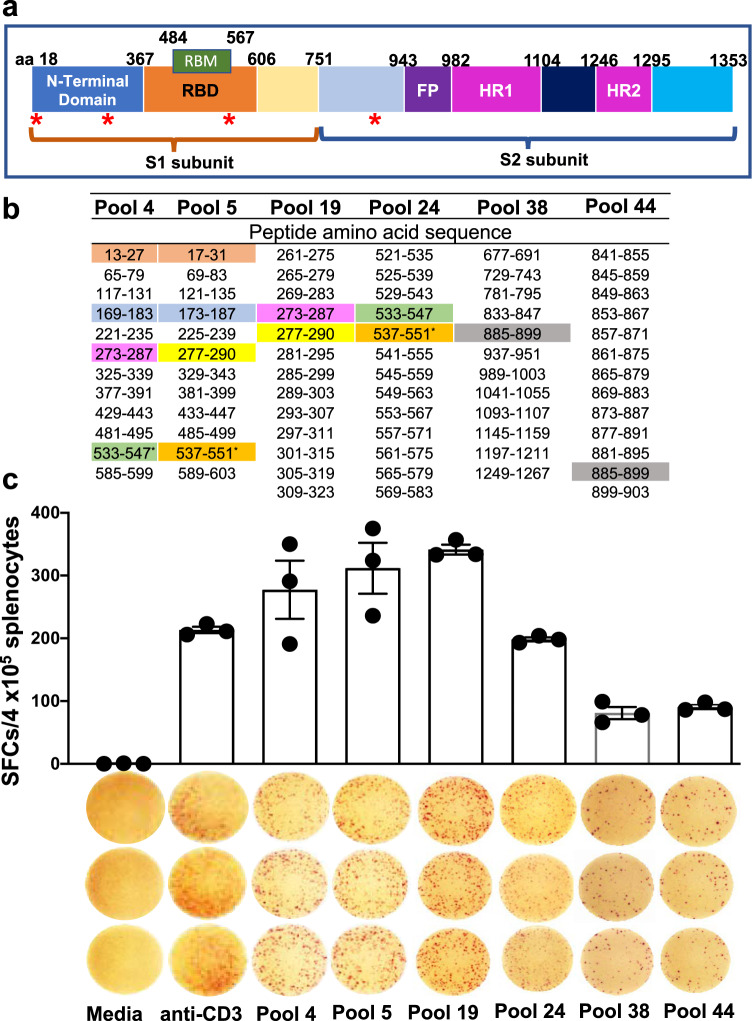
Fig. 6. Epitope mapping data in the splenocytes of SpFN vaccinated mice at week 6 following prime-boost vaccination.
a Schematic representation of the SARS-CoV-2 spike protein showing the different domains with amino acid (aa) numbers. Red asterisk denotes the identified H2Kb/Db-restricted epitopes. b Amino acid residues of SARS-CoV-2 glycoprotein peptide pools. Matrix pools (4, 5, and 38) and overlapping peptide pools of 15-mer peptides overlapping by 11 aa (19, 24, and 48) containing 315 peptides across the entire glycoprotein were tested in the ELISpot assay. Reactivity to specific matching sequences in pools are highlighted with different matching colors and represent the predicted epitopes for H2Kb/Db (iedb.org) or predicted epitopes without ELISpot responses (blue highlight; hypothetical epitope). Amino acid sequences 13–27 (pool 4) and 17–31 (pool 5) have been highlighted in color to indicate confirmed epitopes based on ELISpot reactivity to peptide pool 14. c Number of spike-specific IFN-γ spot forming units stimulated with 52 peptide pools from the Epitope Mapping Peptide Set of the spike protein is presented as scatter plots (mean ± s.d./4 × 105 splenocytes). ELISpot images of triplicate wells and the stimulants are shown on the X-axis. T cell responses were considered positive when mean spot count exceeded the mean ± 3 s.d. of the negative control wells. Splenocytes were stimulated in vitro with media (negative control), anti-CD3 (positive control), or SARS-CoV-2 spike peptide pools 4, 5, 19, 24, 38, and 44.