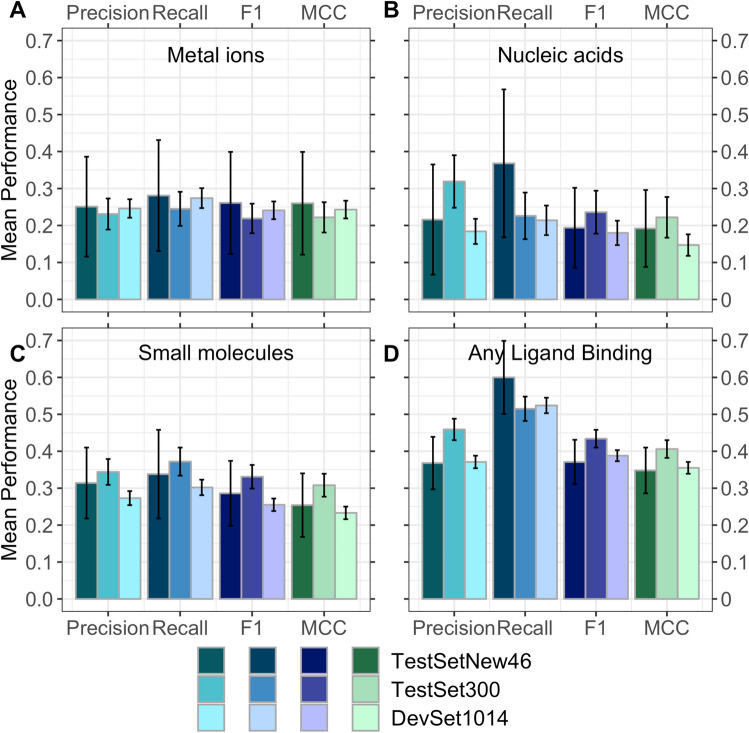
Figure 1.
Performance of new method bindEmbed21DL. Performance captured by four per-residue measures: precision (Eq. 2), recall (Eq. 1), F1 score (Eq. 3), and MCC (Eq. 4). Data sets: DevSet1014 (validation splits of cross-validation development, most light colors), TestSet300 (fixed test set used during development, darker colors), and TestSetNew46 (additional test set compiled after development, most dark colors). Predictions of residues binding to (A) metal ions, (B) nucleic acids (DNA or RNA), (C) small molecules, and (D) any ligand class grouping all three classes into one (considering each residue predicted/observed to bind to one of the three ligand classes as binding, all others as non-binding). On the validation set DevSet1014, bindEmbed21DL predicted any binding residue with F1 = 39 ± 2%. Surprisingly, the number was slightly higher for the test set TestSet300 (F1 = 43 ± 2%) while being similar on the additional test set TestSetNew46 (F1 = 37 ± 6%). Error bars indicate 95% CIs.