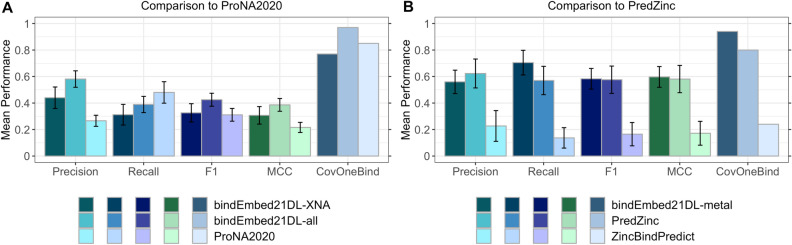
Figure 3.
BindEmbed21DL competitive with specialists. (A) XNA binding. Data: 66 DNA- or RNA-binding (dubbed XNA) proteins from the test set TestSet300. ProNA202019 (lightest shaded bars) uses MSAs to predict DNA-, RNA-, and protein-binding, while the method introduced here uses embeddings only (no MSA); bindEmbed21DL-XNA (darkest shaded bars) marked predictions of either DNA or RNA (XNA); bindEmbed21DL-all (lighter shaded bars) marked using all binding predictions and assessing only XNA-binding. While the difference in F1 scores between the three methods was within the error bars (95% CIs), bindEmbed21DL (-XNA and -all) achieved a statistically significant higher performance than ProNA2020 while ProNA2020 achieved a higher recall. Also, the fraction of proteins with at least one XNA prediction (CovOneBind, Eq. 8) was higher for ProNA2020 than for bindEmbed21DL-XNA. However, when considering any residue predicted as binding (bindEmbed21DL-all: nucleic acid, or metal ion, or small molecule), our new method apparently reached the highest values due to confusions between XNA and other ligands (Supplementary Table S5). (B) Zinc-binding. Data: 51 zinc-binding proteins from TestSet300. ZincBindPredict29 (lightest shaded bars) and PredZinc20 (darker shaded bars) predict zinc-binding; bindEmbed21DL-metal (darkest shaded bars) marked predictions for metal ions. bindEmbed21DL-metal achieved a similar performance as PredZinc, while providing predictions for more proteins (CovOneBind(bindEmbed21DL-metal) = 94% vs. CovOneBind(PredZinc) = 80%). ZincBindPredict was not competitive due to only providing predictions for 12 proteins (CovOneBind(ZincBindPredict) = 24%).