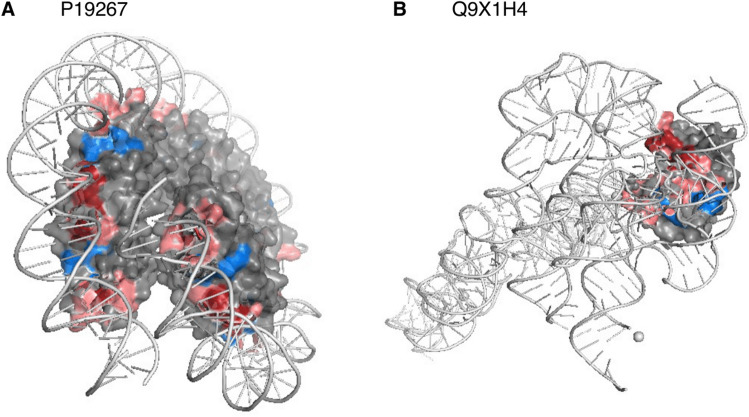
Figure 5.
Annotations from low-resolution structures supported through reliable predictions. (A) Our development set (DevSet1014) contained the PDB structure 1A7W31,32 for the DNA-binding protein HMf-2 (UniProt ID: P19267). No DNA/nucleic acid binding was annotated in that structure, but our new method, bindEmbed21DL, reliably predicted (probability 0.95) four residues to bind nucleic acids. Shown is the PDB structure 5T5K31,33 for the same protein that has a resolution of 4.0 Å and annotations of DNA-binding, including the four most reliable predictions (dark red). Overall, 10 of 13 (77%) residues annotated as DNA-binding in 5T5K were also predicted by bindEmbed21DL (shown in lighter red; blue residues indicate experimental annotations which were not predicted). (B) For the ribonuclease P protein component (UniProt ID: Q9X1H4), four residues were predicted with a probability 0.95 (indicated in dark red), none of these matched the annotations in the PDB structure 6MAX31,34. However, those four residues were considered as binding according to the two low-resolution structures 3Q1Q (3.8 Å)31,35 (visualized) and 3Q1R (4.21 Å)31,35. In total, those structures marked 21 binding residues; 15 of those 21 (71%) were correctly predicted (light red; blue residues observed to bind but not predicted). These two examples highlighted how combining low-resolution experimental data and very reliable predictions from bindEmbed21DL could refine those annotations and/or help designing new investigations.