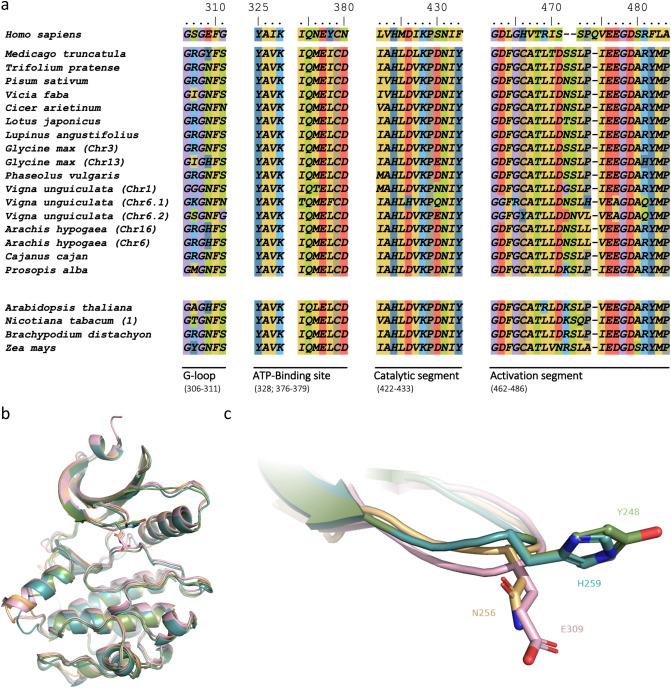
Figure 3.
Sequence alignment of four segments of WEE1 for plants of interest and 3D predictions. (a) Sequences were aligned on MEGAX based on CLUSTAL W algorithm29. Alignments were then edited using BioEdit 7.2.5 software31 (Ibis Biosciences, CA). Here, only four segments are shown: G-loop, ATP-Binding site, Catalytic segment and Activation segment. Amino acid numeration is based on the human (H. sapiens) sequence. Amino acids are colored according to their properties: hydrophobic (AILMFVW) in yellow; acidic (DE) in red; basic (RK) in light blue; polar (QSNT) in green; other aromatic (YH) in dark blue; and C in brown, P in grey and G in purple. Legumes (middle) and model (/examples of dicot and monocot) plants (bottom) are shown. (b) Superimposition of the 3D structure of H. sapiens WEE1 (pink) with predicted ones of A. thaliana (blue), M. truncatula (green) and P. sativum (gold). (c) Detail of the 3D G-loops with the human E309 or its analogs (H259 in Arabidopsis, Y248 in M. truncatula, or N256 in P. sativum) with side chains represented in sticks.