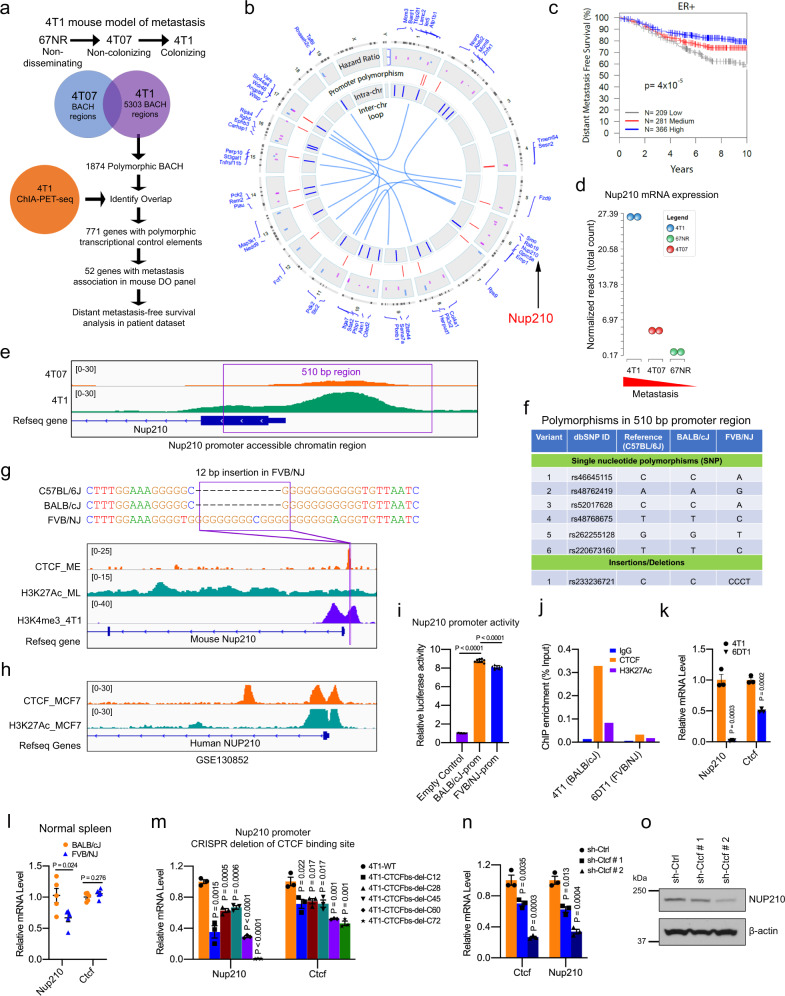
Fig. 1. Genome-wide analysis of noncoding region polymorphisms identifies Nup210 as a metastasis susceptibility gene.
a Scheme for metastasis susceptibility gene identification. b Circos plot representation of the result in (a). Red lines = polymorphic promoter, blue lines = intrachromosomal, and light blue lines = interchromosomal looping interactions. c Distant metastasis-free survival (DMFS) of human ER+ breast cancer patients stratified by their expression of the 52-gene signature. Kaplan–Meier analysis with the log-rank test. d Nup210 mRNA expression in the mouse 4T1 cell line series. e Integrative Genomics Viewer (IGV) track of the Nup210 promoter BACh region in 4T07 and 4T1 cells. f Polymorphisms within the 510 bp Nup210 promoter region. g 12 bp FVB/NJ promoter indel is located within a CTCF-binding site in mouse Nup210 promoter. h CTCF and H3K27Ac enrichment in human NUP210 promoter of MCF7 cells. i Luciferase assay of BALB/cJ and FVB/NJ Nup210 promoter regions. ANOVA, Tukey’s multiple comparison test, mean ± s.e.m, n = 8 biological replicates. j ChIP analysis of CTCF and H3K27Ac at Nup210 promoter of 4T1 and 6DT1 cells. k Nup210 and Ctcf mRNA levels in cell lines (4T1, 6DT1). Two-tailed t test, mean ± s.e.m, n = 3 biological replicates. l Nup210 and Ctcf mRNA levels in mouse spleen (BALB/cJ or FVB/NJ). Two-tailed t test, mean ± s.e.m, n = 5 mice. m Nup210 and Ctcf mRNA level in Nup210 promoter CTCF-binding site-deleted clones. Two-tailed t test, mean ± s.e.m, n = 3 biological replicates. n Effect of Ctcf knockdown on 4T1 Nup210 mRNA levels. Two-tailed t test, mean ± s.e.m, n = 3 biological replicates. o Effect of Ctcf knockdown on NUP210 protein level in 4T1 cells.