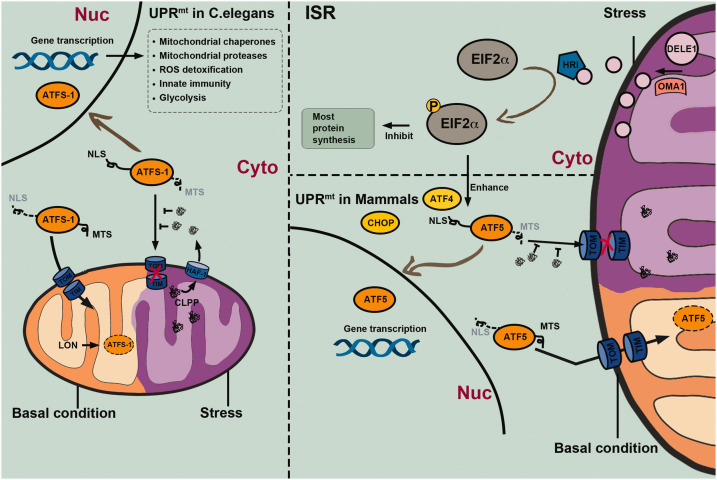
Figure 2.
Mechanisms of the mitochondrial unfolded protein response (UPRmt) in Caenorhabditis elegans and mammalian cells. The UPRmt is an adaptive transcriptional response that can be activated by various mitochondrial stressors. In C. elegans, the key regulator of UPRmt is the activation transcription factor associated with stress 1 (ATFS-1), which is continuously transported into mitochondria via mitochondrial targeting sequence (MTS) guidance and cleaved by the Lon protease under physiological conditions. However, the ATFS-1 mitochondria targeting can be suppressed by the accumulation of aberrant peptides in the cytosol, which are unfolded or misfolded proteins that are cleaved by the caseinolytic protease P (CLPP) and extruded into the cytosol via the transporter HAF-1 under stress conditions. Stabilized ATFS-1 translocates through guidance of the nuclear localization sequence (NLS) into the nucleus from where it promotes gene transcription related to mitochondrial chaperones and proteases, detoxification of reactive oxygen species (ROS), innate immunity, and glycolysis. Together, these protective responses facilitate the recovery of mitochondrial homeostasis. UPRmt in mammalian cells may be regulated by basic leucine zipper (bZip) protein homologs including the C/EBP homologous protein (CHOP), activating transcription factor 4 (ATF4), and activating transcription factor 5 (ATF5), which are transcribed under the control of the integrated stress response (ISR) mechanism mediated by phosphorylated eukaryotic initiation factor-2 alpha subunit (eIF2α). The mitochondrial metallopeptidase OMA1 cleaves the DELE1 protein into short peptides, contributing to heme-regulated inhibitor (HRI) activation, eIF2α phosphorylation, and a general inhibition of gene transcription, while promoting transcription of genes encoding bZip proteins.