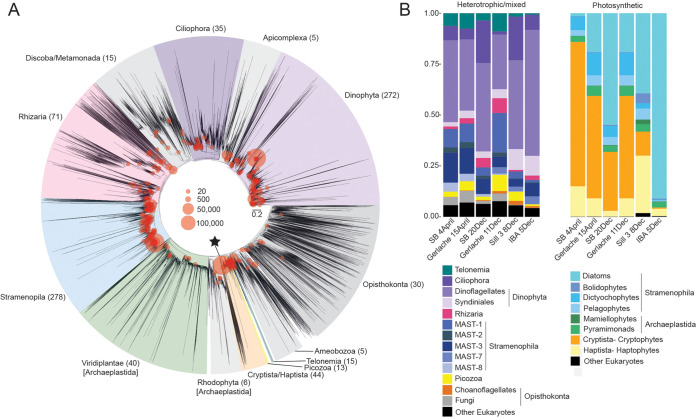
FIG 2.
Eukaryotic diversity and distributions at Andvord Bay and Gerlache Strait stations. (A) Depiction of reference maximum-likelihood phylogenetic construction (based on near-full-length 18S rRNA gene sequences [see Materials and Methods] onto which 18S V9 amplicon sequence variants [ASVs] from the austral spring and fall samples were mapped using published methods [29]). The size of the red circle is proportional to the number of amplicons (within the ASV assigned to the specific node) out of the total number of quality-controlled amplicons generated. Major eukaryotic groups are highlighted, and the number of ASVs represented is given in parentheses next to each group label. The black star indicates the node representing cryptophytes. (B) Relative 18S rRNA gene amplicon frequency for this subset of samples, based on Qiime 2 classification, with a subsequent partitioning based on a literature review of nutritional types. The “heterotrophic/mixed category” includes strict heterotrophs, sequences classified into broad taxonomic groups that include both heterotrophic and mixotrophic (capable of phagotrophic nutrition and photosynthesis) representatives, and groups where the nutritional mode could not be identified (i.e., dinoflagellates). Only groups represented at ≥10% relative contributions in one or more samples are shown. Note that MAST-12 was also present but placed in the “other eukaryotes” classification in panel b, along with other ASVs in taxonomic groups of low relative abundance (<10% in all samples) and ASVs classified as “unidentified” based on Qiime 2 (see Table S2 in the supplemental material). The “other eukaryotes” category under photosynthetic eukaryotes comprised ASVs in taxonomic groups that were either present at low relative abundance (<10% in all samples) or lacked a taxonomic classification beyond “Archaeplastida.” All samples represented were also analyzed using the V1-V2 16S rRNA gene region (see Fig. 4).