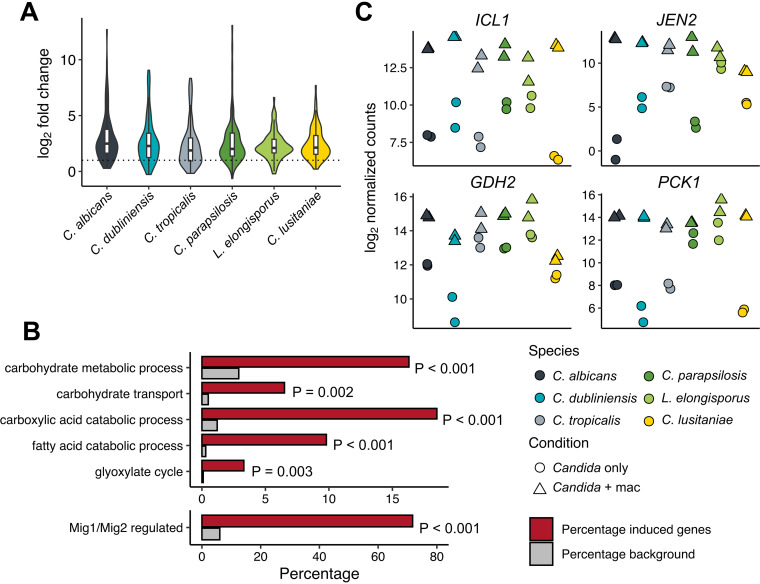
FIG 2.
The core response of CUG clade species primarily consists of alternative carbon metabolism genes. (A) Violin plot of the log2 fold change (LFC) of the 92 genes showing significant induction (FDR = 0.1) across at least five species. Box plots showing median and quartiles are superimposed. The dashed line indicates an LFC of 1 (2-fold induction). (B) Selected enriched GO terms (top) and Mig1/Mig2 regulon inclusion (bottom) among core induced genes depicted in panel A. Analysis was performed using C. albicans orthologs, and the 4,376 conserved genes were the background gene set. FDR-adjusted P values for GO term enrichment are shown. The P value for Mig1/Mig2 regulon inclusion is from a hypergeometric test comparing core induced and total genes. (C) Examples of individual alternative carbon metabolism genes demonstrating conserved phagocytosis-dependent induction.