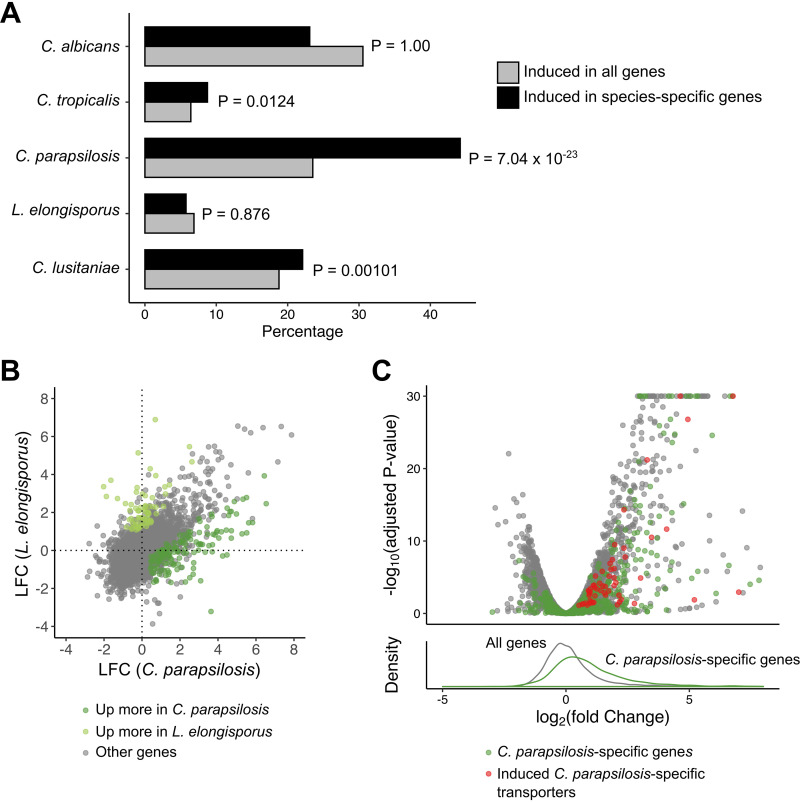
FIG 6.
C. parapsilosis has an enrichment of phagocytosis induction in species-specific genes and enhanced transporter induction compared to related L. elongisporus. (A) For each species, the fraction of unique genes (found only in that species) induced is compared to the fraction of all genes induced in that species. P values are calculated using hypergeometric tests. C. dubliniensis was excluded, as it shares nearly all genes with C. albicans. (B) Comparison of log2 fold change (LFC) between C. parapsilosis and L. elongisporus in response to phagocytosis. (C) Volcano plot of genes in the C. parapsilosis response to phagocytosis (upper panel). C. parapsilosis genes without an ortholog in L. elongisporus are in green (C. parapsilosis-specific induced putative transporter-encoding genes are in red). A histogram (lower panel) of the distribution of LFC for all C. parapsilosis genes (gray) versus C. parapsilosis-specific genes (green) demonstrates an enrichment for the species-specific genes among the induced genes.