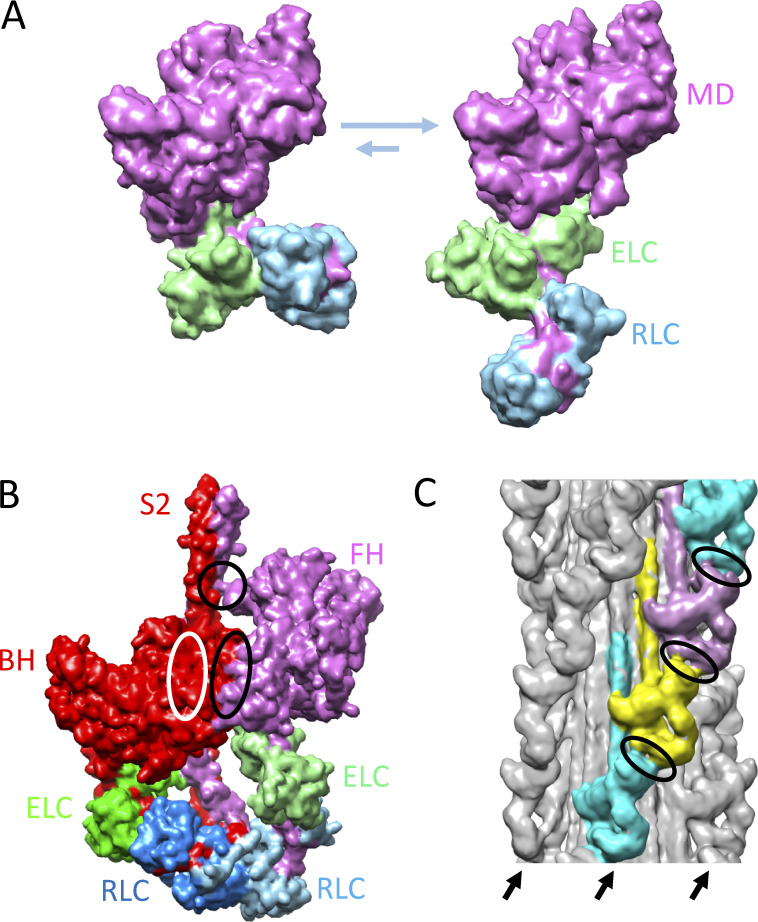
Figure 2.
Structural basis of SRX. (A) Proposed conformations of myosin heads (S1), in equilibrium with each other, underlying SRX (left, bent) and DRX (right, straight) ATP turnover rates (Anderson et al., 2018). MD, motor domain. (B) IHM of cardiac HMM showing BH and FH (based on Protein Data Bank accession no. 5TBY). Ellipses show regions of interaction between BH and FH motor domains (black ellipse), FH and S2 (black circle), and BH and S2 (white ellipse; interaction occurs on rear side of BH). (C) Thick filament (tarantula; EM Data Bank accession no. 1950) showing IHMs lying along four coaxial helices (three on front marked with arrows) creating intermolecular interactions (ellipses) between FH of one IHM and regulatory domain of IHM above. M-line would be at the top of the image. The reconstruction shows the average positions of the heads in the filament; however, the FHs are thought to be dynamic, leaving and returning to the IHM (Fig. 1 B; see text). Models in this figure were created with UCSF Chimera (Pettersen et al, 2004).