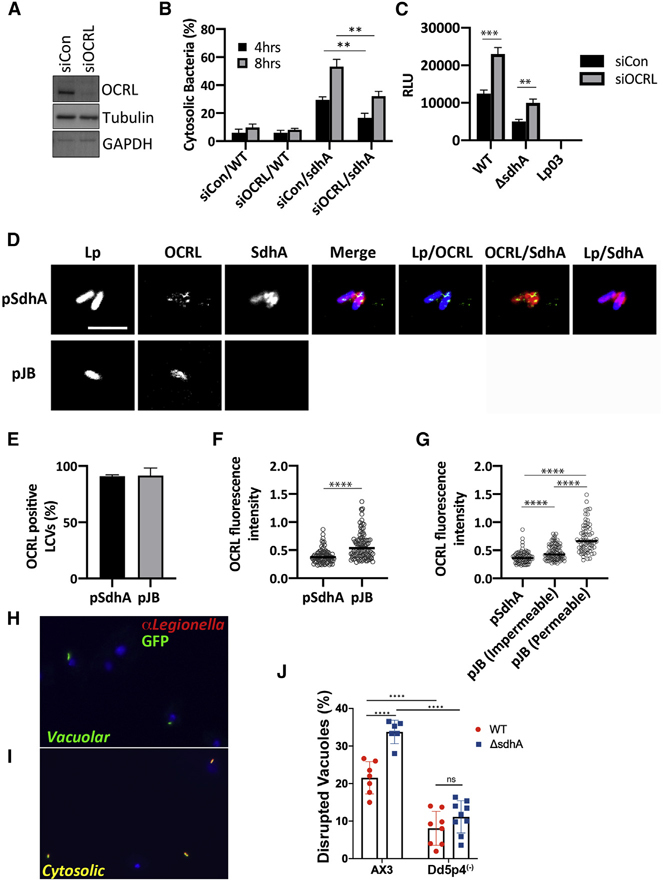
Figure 6. OCRL is linked to vacuole disruption of L. pneumophila sdhA mutants.
(A) COS-7 cells treated with OCRL-targeting (siOCRL) or non-targeting control siRNA (siCon) for 72 h were gel fractionated and immunoprobed with antibodies directed against noted proteins.
(B) COS-7 cells were depleted by siRNA (3 days) and then challenged at MOI = 5 with noted L. pneumophila strains. Cells were fixed and stained for bacteria before and after permeabilization, as in Figure 4B. Data are mean values ± SD of three biological replicates. 100 LCVs per replicate were quantified (**p < 0.01 using two-tailed unpaired Student’s t test).
(C) L. pneumophila lux+ strains were incubated with COS-7 cells at MOI = 20, and bacterial yield was measured 48 h post-infection by relative luminescence (RLU). The replication-deficient dotA null mutant Lp03 was used as a negative control. Data are mean values ± SD of triplicate biological infections (**p < 0.01 and ***p < 0.001 using two-tailed unpaired Student’s t test).
(D) Confocal image showing a section of vacuoles isolated from infected U937 cells (MOI = 10; 3 h) with ΔsdhA mutant harboring pSdhA or pJB vector. The presence of OCRL and SdhA on LCVs was assessed by immunofluorescence using antibodies directed against OCRL, SdhA, and L. pneumophila (scale bar, 4 μm). See also Figure S4.
(E) Quantification of OCRL-positive LCVs containing ΔsdhA mutants with pSdhA or pJB vector at 3 hpi. 85 LCVs were quantified for each infection, and OCRL-positive vacuoles were displayed as percent of total vacuoles analyzed. Data are mean percentage ± SD of biological triplicate infections.
(F) Plot of OCRL intensity associated with LCV, with medians displayed. More than 70 LCVs were quantified per experiment, and data were pooled from 3 experiments (****p < 0.0001; Mann-Whitney U test).
(G) Plot comparing the OCRL intensity on LCVs harboring pSdhA compared to strain harboring empty pJB vector, divided into impermeable and permeable LCVs with medians displayed. More than 70 LCVs were quantified per experiment, and data were pooled from 3 experiments (****p < 0.0001; Mann-Whitney U test).
(H and I) Challenge of D. discoideum strain AX3 (WT) with L. pneumophila-GFP. Fixed cells were probed with α-L. pneumophila followed by secondary (Alexa Fluor 594, red) and analyzed in absence of permeabilization.
(H) Example of vacuolar (intact LCV) bacteria not detected by antibody.
(I) Example of cytosolically exposed bacteria.
(J) D. discoideum AX3 (WT) and Dd5p4 (OCRL−) were challenged for 6 h with either L. pneumophila LP02-GFP (WT) or isogenic ΔsdhA-GFP strains, fixed, and probed with α-L. pneumophila to identify disrupted vacuoles. More than 50 LCVs were imaged per experiment, and disrupted vacuoles were shown as percentage of total number of imaged vacuoles. Biological triplicates were performed with two to three technical replicates each time. Data shown are mean ± SD from all biological and technical replicates pooled to give the individual datapoints displayed (n = 6–9 replicates). Statistical analysis was performed using one-way ANOVA with Tukey’s multiple comparisons. ****p < 0.0001.