Abstract
Non-coding RNAs (ncRNAs) are involved in various cellular processes. There are several ncRNA classes, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). The detailed roles of these molecules in pulmonary hypertension (PH) remain unclear. We systematically collected and reviewed reports describing the functions of ncRNAs (miRNAs, lncRNAs, and circRNAs) in PH through database retrieval and manual literature reading. The characteristics of identified articles, especially the experimental methods, were carefully reviewed. Furthermore, regulatory networks were constructed using ncRNAs and their interacting RNAs or genes. These data were extracted from studies on pulmonary arterial smooth muscle cells, pulmonary artery endothelial cells, and pulmonary artery fibroblasts. We included 14 lncRNAs, 1 circRNA, 74 miRNAs, and 110 mRNAs in the constructed networks. Using these networks, herein, we describe the current knowledge on the role of ncRNAs in PH. Moreover, these networks actively provide an improved understanding of the roles of ncRNAs in PH. The results of this study are crucial for the clinical application of ncRNAs.
Keywords: pulmonary hypertension, long non-coding RNA, circular RNA, microRNA, network
1 Introduction
Pulmonary hypertension (PH) is a serious disease characterized by progressively increased pulmonary vascular resistance and pulmonary artery pressure; the diagnostic criterion is mean pulmonary artery pressure ≥25 mmHg (Galiè et al., 2016; Weber et al., 2018). The increased pulmonary artery pressure in PH results from changes in the structure and function of the vessel wall, which is induced by abnormal pulmonary cell proliferation, apoptosis, and migration (Bourgeois et al., 2018a). Patients with PH may experience dyspnea, fatigue, syncope, chest pain, and/or edema of the legs and ankles. The causes of PH can be broadly classified as primary and secondary causes. To date, ion channels, vasoactive substances, immune factors, and genetic factors are known to be involved in the pathogenesis of PH (Chelladurai et al., 2016; Veith et al., 2016; Bourgeois et al., 2018b).
Recently, many non-coding RNAs (ncRNAs) have been recognized as important regulators in the development of PH. Most human genes (>95%) do not produce proteins but ncRNA molecules. Among them, microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) are the most widely studied. MiRNAs are small ncRNAs containing 21–22 nucleotides, which post-transcriptionally regulate gene expression (Wakiyama and Yokoyama. 2014). LncRNAs, which have more than 200 nucleotides, are transcribed from intergenic or intragenic regions. They can bind to proteins, RNA, or DNA to execute regulatory roles (Botti et al., 2017). CircRNAs are a novel class of ncRNAs with a closed loop structure, making them highly stable and capable of interacting with proteins or RNA (Di et al., 2019). NcRNAs have been identified to regulate multiple steps of gene expression. However, because of the large quantity and diverse mechanisms, it is difficult to comprehensively understand the roles of ncRNAs.
NcRNA-based therapeutics have emerged for several diseases, including PH. An effective ncRNA-based strategy demands a thorough understanding of the diverse and context-dependent regulatory relationships of ncRNAs. The regulation of gene expression by ncRNAs is frequently cell specific, suggesting that not only expression level, but also activity or bioavailability contribute to the biofunction of ncRNAs (Correia de Sousa et al., 2019). Thus, in this article, we reviewed the published literature to search for functional miRNAs, lncRNAs, and circRNAs in PH. Next, we constructed networks of validated ncRNAs and their interacting RNAs or genes to investigate the role of ncRNAs in PH.
2 Screening of Articles
2.1 Criteria for Study Selection
A literature search was performed in PubMed with the query listed in Table 1; we identified 602 articles. In addition, we also reviewed other public databases, including the Human microRNA Disease Database v3.2, miRWalk 2.0, and LncRNADisease v2.0, to identify validated functional ncRNAs in PH. Studies were selected when the following criteria were met: 1) the study reported pathogenic roles of miRNAs, lncRNAs, and/or circRNAs in PH; 2) mechanistic studies were performed in pulmonary arterial smooth muscle cells (PASMCs), pulmonary artery endothelial cells (PAECs), and/or pulmonary artery fibroblasts (PAFs); and 3) the relationships between ncRNAs and their interacting RNAs or genes were experimentally identified via luciferase reporter assay, western blot, and/or qPCR. Using these criteria returned 140 qualified articles (Figure 1).
TABLE 1.
Query for searching articles from PubMed.
| Query | Number of articles |
|---|---|
| (“rna, untranslated"[MeSH Terms] or “non-coding RNA” or “ncRNA” or “noncoding RNA” or “RNA, Long Noncoding"[Mesh] or “long non-coding RNA” or “lncRNA” or “long intergenic non-coding RNA” or “lincRNA” or “RNA, Circular"[Mesh] or “circRNA” or “circular RNA” or “MicroRNAs"[Mesh] or “microRNA” OR “miRNA”) and (“PAH” or “pulmonary hypertension” or “pulmonary artery hypertension") | 602 |
FIGURE 1.
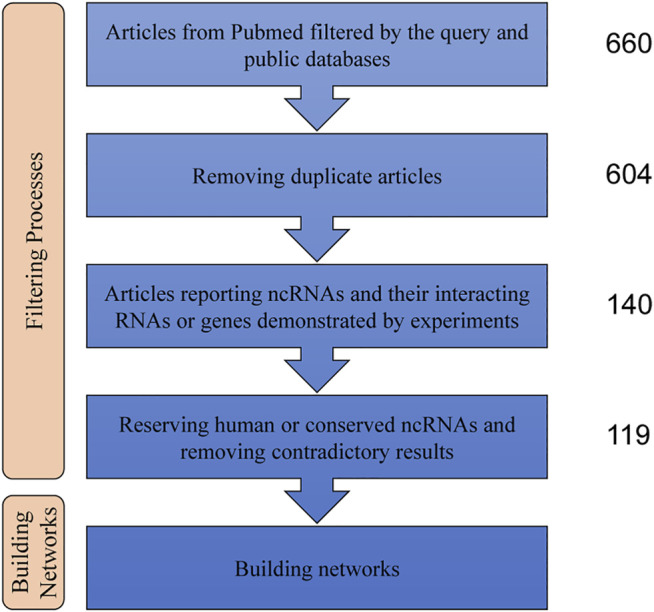
Steps of the data analysis used to build the ncRNA networks. NcRNAs: non-coding RNAs.
2.2 General Characteristics of Qualified Articles
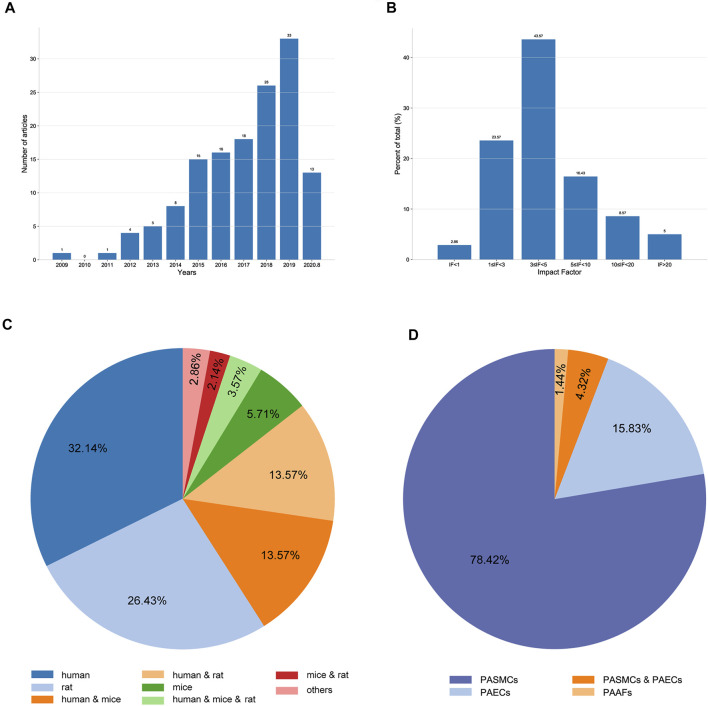
When sorted by publication date, we found that the number of eligible articles continuously increased year by year (Figure 2A). The impact factors (IF) of the articles ranged from 0 to 36.13; articles with 3 ≤ IF < 5 accounted for the highest proportion (Figure 2B). Of the 140 qualified articles, 32.14% were studies using human tissues or cells. In studies using experimental animals, rats were the most commonly used, accounting for 26.43% of the total studies (Figure 2C). Moreover, when classified by cell type, 78.42, 15.83, 1.44, and 4.32% of studies were performed in PASMCs, PAECs, PAFs, and both PASMCs and PAECs, respectively (Figure 2D).
FIGURE 2.
Characteristics of the extracted studies. (A) Distribution of the included articles according to the publication year. (B) Distribution of the included articles according to the impact factor. (C) Proportions of different species studied in the included articles. (D) Proportions of different cell types used in the included articles.
3 Non-coding RNA Networks for Pulmonary Hypertension
3.1 Construction of Non-coding RNA Regulatory Networks
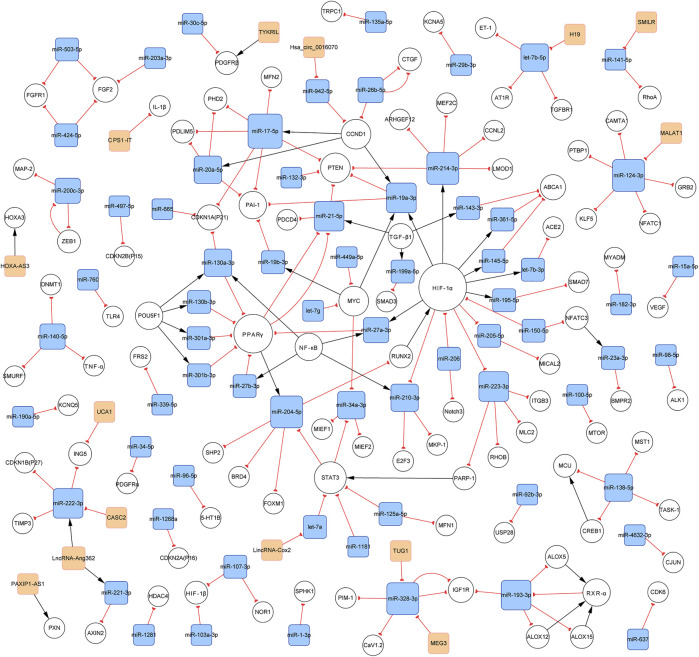
Regulatory networks were constructed using ncRNAs and their interacting RNAs or genes in PASMCs, PAECs, and PAFs. Given ncRNA conservation among species, only human ncRNAs or ncRNAs that were conserved between human and experimental animals were included. If there were contradictory results, the results from higher-impact articles were selected. In addition, some crucial regulatory relationships between protein-coding genes and validated transcription factor–miRNA interactions from TransmiR v2.0 were also described in the networks to present an in-depth explanation on the roles of ncRNAs in PH. The nodes represented interacting molecules, and the edges represented the regulatory connections. Each edge indicated a publication supporting the connection. Square and circular nodes represented ncRNAs and coding RNAs or genes, respectively. Node color was based on the type of molecule (lncRNAs and circRNAs are orange, miRNAs are blue, and coding RNAs or genes are empty). Node sizes represented their degrees (number of edges that directly link to the node). Edges represented the regulatory connections: red edges depicted links indicating repressive action (semicircular arrow heads), and black edges indicated activation (traditional arrow heads). The nodes in this network were involved in cell proliferation, apoptosis, migration, metabolism, endothelial–mesenchymal transition, and extracellular matrix remodeling. The steps used in our approach are shown in Figure 1.
3.2 General Characteristics of the Constructed Networks
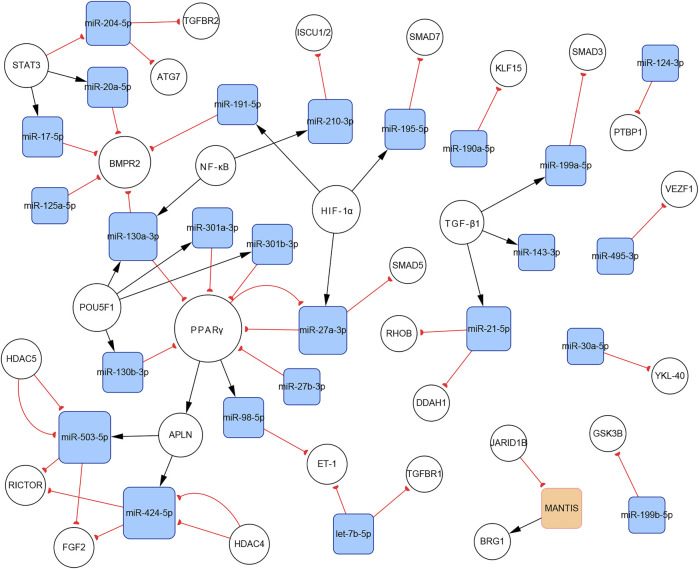
In total, 140 articles describing 14 lncRNAs, 1 circRNA, 74 miRNAs, and 110 mRNAs, were included in our networks. Considering the unique biological characteristics of different cell types, we constructed networks according to cell type. The network of PASMCs contained 13 lncRNAs, 1 circRNA, 69 miRNAs, and 96 mRNAs. The network of PAECs contained 1 lncRNA, 25 miRNAs, and 29 mRNAs. The network of PAFs contained 6 miRNAs and four mRNAs. The networks are shown in Figures 3–5. Detailed network compositions are listed in Table 2.
FIGURE 3.
PH-associated network of ncRNAs and their interacting RNAs or genes in PASMCs. The square and circular nodes represent ncRNAs and coding RNAs or genes, respectively. Node color is based on the type of molecule (lncRNAs and circRNAs are orange, miRNAs are blue, and coding RNAs or genes are empty). Node sizes represent the degrees (the number of edges that directly link to the node). Edges represent the regulatory connections, and each edge indicates a publication. When multiple publications describe one interaction, multiple edges connect the same two nodes. Red edges depict links indicating repressive action (semicircular arrow heads), and black edges are those indicating activation (with traditional arrow heads). The nodes in this network are involved in cell proliferation, apoptosis, migration, and metabolism. PH: pulmonary hypertension; PASMCs: pulmonary artery smooth muscle cells; lncRNAs: long non-coding RNAs; circRNA: circular RNA; miRNA: microRNA.
FIGURE 5.
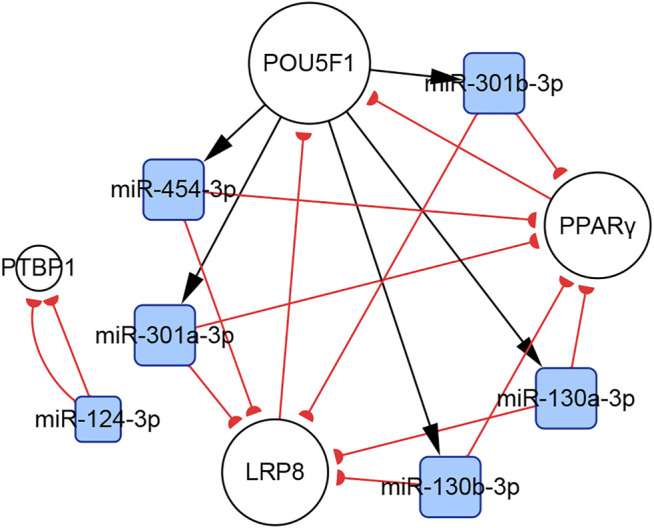
The PH-associated network of ncRNAs and their interacting RNAs or genes in PAFs. The square and circular nodes represent ncRNAs and coding RNAs or genes, respectively. Node color is based on the type of molecule (miRNAs are blue, coding RNAs or genes are empty). Node sizes represent the degrees (number of edges that directly link to the node). Edges represent regulatory connections. Each edge indicates a publication. When multiple publications describe one interaction, multiple edges connect the same two nodes. Red edges depict links indicating repressive action (semicircular arrow heads), and black edges are those indicating activation (traditional arrow heads). The nodes in this network were primarily involved in cell proliferation and extracellular matrix remodeling. PAFs: pulmonary artery fibroblasts.
TABLE 2.
List of network interactions.
| Upstream molecule | Downstream molecule | Interaction type a | PMID | Reference |
|---|---|---|---|---|
| ALOX12 | RXR-α | pos | 24963038 | Sharma et al. (2014) |
| ALOX15 | RXR-α | pos | 24963038 | Sharma et al. (2014) |
| ALOX5 | RXR-α | pos | 24963038 | Sharma et al. (2014) |
| APLN | miR-424-5p | pos | 23263626 | Kim et al. (2013) |
| APLN | miR-503-5p | pos | 23263626 | Kim et al. (2013) |
| CASC2 | miR-222-3p | neg | 32206065 | Han et al. (2020) |
| CCND1 | miR-17-5p | pos | 18695042 | Yu et al. (2008) |
| CCND1 | miR-19a-3p | pos | 28090171 | Inoue and Fry. (2015) |
| CCND1 | miR-20a-5p | Pos | 28090171 | Inoue and Fry. (2015) |
| CPS1-IT | IL-1β | neg | 30982984 | Zhang et al. (2019b) |
| CREB1 | MCU | pos | 27648837 | Hong et al. (2017) |
| H19 | let-7b-5p | neg | 30547791 | Su et al. (2018) |
| HDAC4 | miR-424-5p | neg | 29102771 | Takagi et al. (2018) |
| HDAC4 | miR-503-5p | neg | 29102771 | Takagi et al. (2018) |
| HDAC5 | miR-424-5p | neg | 29102771 | Takagi et al. (2018) |
| HDAC5 | miR-503-5p | neg | 29102771 | Takagi et al. (2018) |
| HIF-1α | let-7b-3p | pos | 30628484 | Zhang H et al. (2019) |
| HIF-1α | miR-145-5p | pos | 25129238 | Agrawal et al. (2014) |
| HIF-1α | miR-191-5p | pos | 25119596 | Song et al. (2014) |
| HIF-1α | miR-195-5p | pos | 28862358 | Zeng et al. (2018) |
| HIF-1α | miR-19a-3p | pos | 31682848 | Zhao et al. (2019) |
| HIF-1α | miR-205-5p | pos | 23924028 | Gandellini et al. (2014) |
| HIF-1α | miR-210-3p | neg | 22886504 | Gou et al. (2012) |
| HIF-1α | miR-214-3p | pos | 24011070 | el Azzouzi et al. (2013) |
| HIF-1α | miR-223-3p | neg | 26084306 | Meloche et al. (2015a) |
| HIF-1α | miR-27a-3p | pos | 24517586 | Camps et al. (2014) |
| HIF-1α | miR-361-5p | pos | 29339076 | Zhang Y et al. (2018) |
| HOXA-AS3 | HOXA3 | pos | 30304383 | Zhang R et al. (2019) |
| Hsa_circ_0016070 | miR-942-5p | neg | 31593832 | Zhou et al. (2019) |
| JARID1B | MANTIS | neg | 2,8351900 | Leisegang et al. (2017) |
| let-7a | STAT3 | neg | 32803651 | Cheng et al. (2020) |
| let-7b-3p | ACE2 | neg | 30628484 | Zhang Y et al. (2019) |
| let-7b-5p | AT1R | neg | 30547791 | Su et al. (2018) |
| let-7b-5p | ET-1 | neg | 24978044 | Guo et al. (2014) |
| let-7b-5p | TGFBR1 | neg | 24978044 | Guo et al. (2014) |
| let-7g | MYC | neg | 27889560 | Zhang W.-F et al. (2017) |
| LincRNA-Cox2 | let-7a | neg | 32803651 | Cheng et al. (2020) |
| LncRNA-Ang362 | miR-221-3p | pos | 31313741 | Wang et al. (2020) |
| LncRNA-Ang362 | miR-222-3p | pos | 31313741 | Wang et al. (2020) |
| LRP8 | POU5F1 | neg | 26565914 | Bertero et al. (2015) |
| MALAT1 | miR-124-3p | neg | 31257528 | Wang S et al. (2019) |
| MANTIS | BRG1 | pos | 2,8351900 | Leisegang et al. (2017) |
| MEG3 | miR-328-3p | neg | 31477557 | Xing X.-Q et al. (2019) |
| miR-100-5p | MTOR | neg | 26409044 | Wang et al. (2015) |
| miR-103a-3p | HIF-1β | neg | 26827991 | Deng et al. (2016) |
| miR-107-3p | HIF-1β | neg | 26827991 | Deng et al. (2016) |
| miR-107-3p | NOR1 | neg | 31933977 | Chen et al. (2019) |
| miR-1181 | STAT3 | neg | 30211651 | Qian et al. (2018) |
| miR-124-3p | CAMTA1 | neg | 23853098 | Kang K et al. (2013) |
| miR-124-3p | GRB2 | neg | 28496318 | Li Y et al. (2017) |
| miR-124-3p | KLF5 | neg | 31257528 | Wang D et al. (2019) |
| miR-124-3p | NFATC1 | neg | 23853098 | Kang B.-Y et al. (2013) |
| miR-124-3p | PTBP1 | neg | 23853098 | Kang K et al. (2013) |
| miR-124-3p | PTBP1 | neg | 24122720 | Wang et al. (2014) |
| miR-124-3p | PTBP1 | neg | 2,8971999 | Caruso et al. (2017) |
| miR-124-3p | PTBP1 | neg | 2,8972001 | Zhang H et al. (2017) |
| miR-125a-5p | BMPR2 | neg | 25854878 | Huber et al. (2015) |
| miR-125a-5p | MFN1 | neg | 28593577 | Ma et al. (2017) |
| miR-125a-5p | STAT3 | neg | 29700287 | Cai et al. (2018) |
| miR-1268a | CDKN2A(P16) | neg | 31370272 | Lee and Kang. (2019) |
| miR-1281 | HDAC4 | neg | 29514810 | Li et al. (2018) |
| miR-130a-3p | BMPR2 | neg | 28755990 | Li L et al. (2017) |
| miR-130a-3p | CDKN1A(P21) | neg | 25681685 | Brock et al. (2015) |
| miR-130a-3p | LRP8 | neg | 26565914 | Bertero et al. (2015) |
| miR-130a-3p | PPARγ | neg | 24960162 | Bertero et al. (2014) |
| miR-130a-3p | PPARγ | neg | 26565914 | Bertero et al. (2015) |
| miR-130b-3p | LRP8 | neg | 26565914 | Bertero et al. (2015) |
| miR-130b-3p | PPARγ | neg | 24960162 | Bertero et al. (2014) |
| miR-130b-3p | PPARγ | neg | 26565914 | Bertero et al. (2015) |
| miR-132-3p | PTEN | neg | 30896881 | Zeng et al. (2019) |
| miR-135a-5p | TRPC1 | neg | 30038339 | Liu A et al. (2019) |
| miR-138-5p | CREB1 | neg | 27648837 | Hong et al. (2017) |
| miR-138-5p | MCU | neg | 27648837 | Hong et al. (2017) |
| miR-138-5p | MST1 | neg | 23485012 | Li et al. (2013) |
| miR-138-5p | TASK-1 | neg | 29257242 | Liu G et al. (2018) |
| miR-1-3p | SPHK1 | neg | 29167124 | Sysol et al. (2018) |
| miR-140-5p | DNMT1 | neg | 27021683 | Zhang and Xu. (2016) |
| miR-140-5p | SMURF1 | neg | 27214554 | Rothman et al. (2016) |
| miR-140-5p | TNF-α | neg | 30367500 | Zhu et al. (2019b) |
| miR-143-3p | ABCA1 | neg | 30195228 | Yue et al. (2018) |
| miR-141-5p | RHOA | neg | 32559140 | Lei et al. (2020) |
| miR-145-5p | ABCA1 | neg | 30195228 | Yue et al. (2018) |
| miR-150-5p | HIF-1α | neg | 28715868 | Chen M et al. (2017) |
| miR-150-5p | NFATC3 | neg | 30551428 | Li et al. (2019) |
| miR-15a-5p | VEGF | neg | 31894295 | Zhang et al. (2020) |
| miR-17-5p | BMPR2 | neg | 19390056 | Brock et al. (2009) |
| miR-17-5p | CDKN1A(P21) | neg | 30305109 | Liu J. J et al. (2018) |
| miR-17-5p | MFN2 | neg | 27640178 | Lu et al. (2016) |
| miR-17-5p | PAI-1 | neg | 29644896 | Chen K.-H et al. (2018) |
| miR-17-5p | PDLIM5 | neg | 25647182 | Chen et al. (2015) |
| miR-17-5p | PHD2 | neg | 27919930 | Chen et al. (2016) |
| miR-17-5p | PTEN | neg | 30305109 | Liu G et al. (2018) |
| miR-182-3p | MYADM | neg | 32373233 | Sun et al. (2020) |
| miR-190a-5p | KCNQ5 | neg | 24446351 | Li et al. (2014) |
| miR-190a-5p | KLF15 | neg | 30538440 | Jiang et al. (2018) |
| miR-191-5p | BMPR2 | neg | 31119161 | Zhang Z et al. (2019) |
| miR-193-3p | ALOX12 | neg | 24963038 | Sharma et al. (2014) |
| miR-193-3p | ALOX15 | neg | 24963038 | Sharma et al. (2014) |
| miR-193-3p | ALOX5 | neg | 24963038 | Sharma et al. (2014) |
| miR-193-3p | IGF1R | neg | 24963038 | Sharma et al. (2014) |
| miR-195-5p | SMAD7 | neg | 28862358 | Zeng et al. (2018) |
| miR-199a-5p | SMAD3 | neg | 27038547 | Liu H et al. (2016) |
| miR-199b-5p | GSK3B | neg | 27188753 | Wu et al. (2016) |
| miR-19a-3p | PAI-1 | neg | 29644896 | Chen T et al. (2018) |
| miR-19a-3p | PTEN | neg | 31682848 | Zhao et al. (2019) |
| miR-19b-3p | PAI-1 | neg | 29644896 | Chen K.-H et al. (2018) |
| miR-200c-3p | MAP2 | neg | 29044995 | Yuan et al. (2017) |
| miR-200c-3p | ZEB1 | neg | 29044995 | Yuan et al. (2017) |
| miR-203a-3p | FGF2 | neg | 30575929 | Wang et al. (2018) |
| miR-204-5p | ATG7 | neg | 31542480 | Liu H.-M et al. (2019) |
| miR-204-5p | BRD4 | neg | 26224795 | Meloche et al. (2015a) |
| miR-204-5p | FOXM1 | neg | 29290032 | Bourgeois et al. (2018b) |
| miR-204-5p | RUNX2 | neg | 27149112 | Ruffenach et al. (2016) |
| miR-204-5p | SHP2 | neg | 21321078 | Courboulin et al. (2011) |
| miR-204-5p | TGFBR2 | neg | 29196166 | Yu et al. (2018) |
| miR-205-5p | MICAL2 | neg | 30853343 | Tao et al. (2019) |
| miR-206 | Notch3 | neg | 23071643 | Jalali et al. (2012) |
| miR-206 | HIF-1α | neg | 23628900 | Yue et al. (2013) |
| miR-20a-5p | BMPR2 | neg | 19390056 | Brock et al. (2009) |
| miR-20a-5p | PAI-1 | neg | 29644896 | Chen T et al. (2018) |
| miR-20a-5p | PDLIM5 | neg | 25647182 | Chen et al. (2015) |
| miR-20a-5p | PHD2 | neg | 27919930 | Chen et al. (2016) |
| miR-210-3p | E2F3 | neg | 22886504 | Gou et al. (2012) |
| miR-210-3p | ISCU1/2 | neg | 25825391 | White et al. (2015) |
| miR-210-3p | MKP-1 | neg | 25044272 | Jin et al. (2015) |
| miR-214-3p | ARHGEF12 | neg | 31373336 | Xing Y et al. (2019) |
| miR-214-3p | CCNL2 | neg | 27381447 | Liu Y et al. (2016) |
| miR-214-3p | LMOD1 | neg | 27144530 | Sahoo et al. (2016) |
| miR-214-3p | MEF2C | neg | 27144530 | Sahoo et al. (2016) |
| miR-214-3p | PTEN | neg | 28684904 | Liu et al. (2017) |
| miR-21-5p | DDAH1 | neg | 24895913 | Iannone et al. (2014) |
| miR-21-5p | PDCD4 | neg | 28522568 | Green et al. (2017) |
| miR-21-5p | PTEN | neg | 26208095 | Green et al. (2015) |
| miR-21-5p | RHOB | neg | 22371328 | Parikh et al. (2012) |
| miR-221-3p | AXIN2 | neg | 28694128 | Nie et al. (2019) |
| miR-222-3p | ING5 | neg | 32206065 | Han et al. (2020) |
| miR-222-3p | CDKN1B(P27) | neg | 28854428 | Xu et al. (2017) |
| miR-222-3p | TIMP3 | neg | 28854428 | Xu et al. (2017) |
| miR-223-3p | ITGB3 | neg | 30507047 | Liu et al. (2019a) |
| miR-223-3p | MLC2 | neg | 27121304 | Zeng et al. (2016) |
| miR-223-3p | PARP1 | neg | 26084306 | Meloche et al. (2015b) |
| miR-223-3p | RHOB | neg | 27121304 | Zeng et al. (2016) |
| miR-23a-3p | BMPR2 | neg | 29864909 | Zhang X et al. (2018) |
| miR-26b-5p | CCND1 | neg | 2,7322082 | Wang P et al. (2016) |
| miR-26b-5p | CTGF | neg | 2,7322082 | Wang R et al. (2016) |
| miR-26b-5p | CTGF | neg | 28816418 | Zhou et al. (2018) |
| miR-27a-3p | PPARγ | neg | 24244514 | Kang B.-Y et al. (2013) |
| miR-27a-3p | PPARγ | neg | 28484848 | Xie et al. (2017) |
| miR-27a-3p | SMAD5 | neg | 31004656 | Liu et al. (2019b) |
| miR-27b-3p | PPARγ | neg | 25795136 | Bi et al. (2015) |
| miR-27b-3p | PPARγ | neg | 28484848 | Xie et al. (2017) |
| miR-29b-3p | KCNA5 | neg | 31553627 | Babicheva et al. (2020) |
| miR-301a-3p | LRP8 | neg | 26565914 | Bertero et al. (2015) |
| miR-301a-3p | PPARγ | neg | 24960162 | Bertero et al. (2014) |
| miR-301a-3p | PPARγ | neg | 26565914 | Bertero et al. (2015) |
| miR-301b-3p | LRP8 | neg | 26565914 | Bertero et al. (2015) |
| miR-301b-3p | PPARγ | neg | 24960162 | Bertero et al. (2014) |
| miR-301b-3p | PPARγ | neg | 26565914 | Bertero et al. (2015) |
| miR-30a-5p | YKL-40 | neg | 31115541 | Tan et al. (2019) |
| miR-30c-5p | PDGFRβ | neg | 25882492 | Xing et al. (2015) |
| miR-328-3p | CaV1.2 | neg | 22392900 | Guo et al. (2012) |
| miR-328-3p | IGF1R | neg | 22392900 | Guo et al. (2012) |
| miR-328-3p | IGF1R | neg | 31477557 | Xing X.-Q et al. (2019) |
| miR-328-3p | PIM-1 | neg | 27448984 | Qian et al. (2016) |
| miR-339-5p | FRS2 | neg | 28947594 | Chen J et al. (2017) |
| miR-34-5p | PDGFRα | neg | 27302634 | Wang P et al. (2016) |
| miR-34a-3p | MIEF1 | neg | 29431643 | Chen K.-H et al. (2018) |
| miR-34a-3p | MIEF2 | neg | 29431643 | Chen T et al. (2018) |
| miR-361-5p | ABCA1 | neg | 29339076 | Zhang Y et al. (2018) |
| miR-424-5p | FGF2 | neg | 23263626 | Kim et al. (2013) |
| miR-424-5p | FGF2 | neg | 24960162 | Bertero et al. (2014) |
| miR-424-5p | FGFR1 | neg | 23263626 | Kim et al. (2013) |
| miR-424-5p | RICTOR | neg | 29102771 | Takagi et al. (2018) |
| miR-449a-5p | MYC | neg | 30715622 | Zhang et al. (2019a) |
| miR-454-3p | LRP8 | neg | 26565914 | Bertero et al. (2015) |
| miR-454-3p | PPARγ | neg | 26565914 | Bertero et al. (2015) |
| miR-4632-3p | CJUN | neg | 28701355 | Qian et al. (2017) |
| miR-495-3p | VEZF1 | neg | 31030195 | Fu et al. (2019) |
| miR-497-5p | CDKN2B(P15) | neg | 31370272 | Lee and Kang. (2019) |
| miR-503-5p | FGF2 | neg | 23263626 | Kim et al. (2013) |
| miR-503-5p | FGF2 | neg | 24960162 | Bertero et al. (2014) |
| miR-503-5p | FGFR1 | neg | 23263626 | Kim et al. (2013) |
| miR-503-5p | RICTOR | neg | 29102771 | Takagi et al. (2018) |
| miR-637 | CDK6 | neg | 27794186 | Sang et al. (2016) |
| miR-665 | CDKN1A(P21) | neg | 31370272 | Lee and Kang. (2019) |
| miR-760 | TLR4 | neg | 30226538 | Yang et al. (2018) |
| miR-92b-3p | USP28 | neg | 30149918 | Hao et al. (2018) |
| miR-942-5p | CCND1 | neg | 31593832 | Zhou et al. (2019) |
| miR-96-5p | 5-HT1B | neg | 25871906 | Wallace et al. (2015) |
| miR-98-5p | ALK1 | neg | 31322216 | Li et al. (2019) |
| miR-98-5p | ET-1 | neg | 26098770 | Kang et al. (2016) |
| MYC | miR-19a-3p | pos | 17943719 | Schulte et al. (2008) |
| MYC | miR-19b-3p | pos | 17943719 | Schulte et al. (2008) |
| MYC | miR-34a-3p | neg | 18066065 | Chang et al. (2008) |
| NFATC3 | miR-23a-3p | pos | 19574461 | Lin et al. (2009) |
| NF-κB | miR-130a-3p | pos | 28755990 | Li Q et al. (2017) |
| NF-κB | miR-210-3p | pos | 25341039 | Liu et al. (2014) |
| NF-κB | miR-27a-3p | pos | 28484848 | Xie et al. (2017) |
| NF-κB | miR-27b-3p | pos | 28484848 | Xie et al. (2017) |
| PARP-1 | STAT3 | pos | 24270264 | Meloche et al. (2014) |
| PAXIP1-AS1 | PXN | pos | 30450722 | Jandl et al. (2019) |
| POU5F1 | miR-130a-3p | pos | 24960162 | Bertero et al. (2014) |
| POU5F1 | miR-130a-3p | pos | 26565914 | Bertero et al. (2015) |
| POU5F1 | miR-130b-3p | pos | 24960162 | Bertero et al. (2014) |
| POU5F1 | miR-130b-3p | pos | 26565914 | Bertero et al. (2015) |
| POU5F1 | miR-301a-3p | pos | 24960162 | Bertero et al. (2014) |
| POU5F1 | miR-301a-3p | pos | 26565914 | Bertero et al. (2015) |
| POU5F1 | miR-301b-3p | pos | 24960162 | Bertero et al. (2014) |
| POU5F1 | miR-301b-3p | pos | 26565914 | Bertero et al. (2015) |
| POU5F1 | miR-454-3p | pos | 26565914 | Bertero et al. (2015) |
| PPARγ | APLN | pos | 24960162 | Bertero et al. (2014) |
| PPARγ | miR-204-5p | pos | 24960162 | Bertero et al. (2014) |
| PPARγ | miR-21-5p | neg | 26208095 | Green et al. (2015) |
| PPARγ | miR-21-5p | neg | 28522568 | Green et al. (2017) |
| PPARγ | miR-27a-3p | neg | 24244514 | Kang K et al. (2013) |
| PPARγ | miR-98-5p | pos | 26098770 | Kang et al. (2016) |
| PPARγ | POU5F1 | neg | 26565914 | Bertero et al. (2015) |
| RUNX2 | HIF-1α | pos | 27149112 | Ruffenach et al. (2016) |
| RXR-α | miR-193-3p | neg | 24963038 | Sharma et al. (2014) |
| SMILR | miR-141-5p | neg | 32559140 | Lei et al. (2020) |
| STAT3 | miR-17-5p | pos | 19390056 | Brock et al. (2009) |
| STAT3 | miR-204-5p | neg | 23975026 | Xu et al. (2013) |
| STAT3 | miR-20a-5p | pos | 19390056 | Brock et al. (2009) |
| STAT3 | miR-34a-3p | neg | 24642471 | Rokavec et al. (2014) |
| TGF-β1 | miR-143-3p | pos | 2,6311719 | Deng et al. (2015) |
| TGF-β1 | miR-199a-5p | pos | 20705240 | Davis et al. (2010) |
| TGF-β1 | miR-21-5p | pos | 20705240 | Davis et al. (2010) |
| TUG1 | miR-328-3p | neg | 31679623 | Wang D et al. (2019) |
| TYKRIL | PDGFRβ | pos | 32634060 | Zehendner et al. (2020) |
| UCA1 | ING5 | neg | 30353369 | Zhu et al. (2019a) |
| ZEB1 | miR-200c-3p | neg | 18829540 | Bracken et al. (2008) |
pos: positive interaction, neg: negative interaction.
FIGURE 4.
The PH-associated network of ncRNAs and their interacting RNAs or genes in PAECs. The square and circular nodes represent ncRNAs and coding RNAs or genes, respectively. Node color is based on the type of molecule (lncRNAs are orange, miRNAs are blue, and coding RNAs or genes are empty). Node sizes represent the degrees (number of edges that directly link to the node). Edges represent regulatory connections. Each edge indicates a publication. When multiple publications describe one interaction, multiple edges connect the same two nodes. Red edges depict links indicating repressive action (semicircular arrow heads), and black edges represent those indicating activation (traditional arrow heads). The nodes in this network were primarily involved in proliferation, apoptosis resistance, migration, and endothelial–mesenchymal transition. PAECs: pulmonary artery endothelial cells.
3.3 Functional Enrichment Analysis
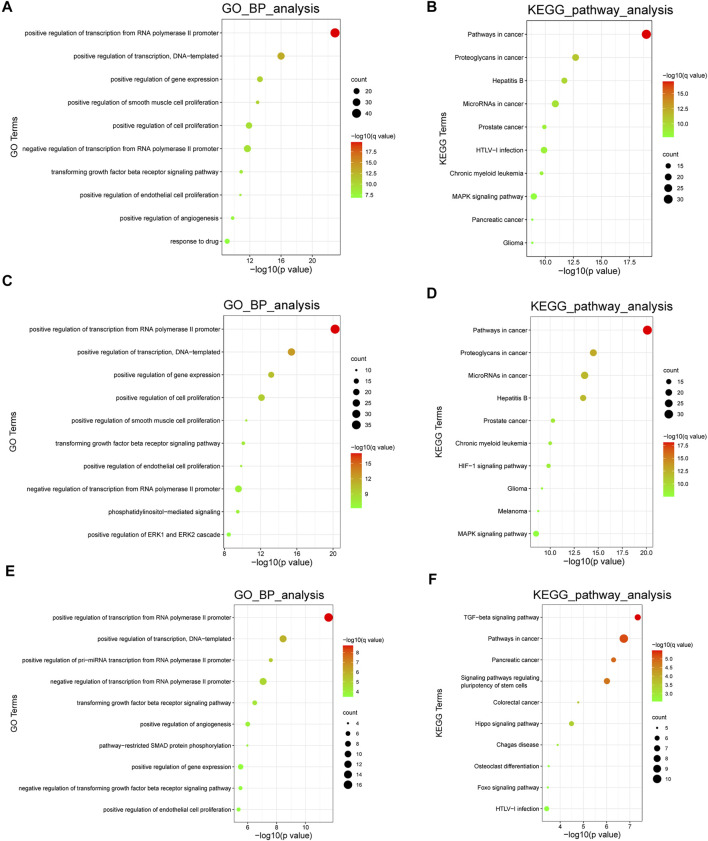
We performed gene ontology (GO) biological process term analyses and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses using the database for Annotation, Visualization, and Integration Discovery (DAVID). The calculation process was dependent on a hypergeometric test, using a statistical significance threshold of p < 0.05 with a false discovery rate (FDR) correction. These analyses provided a general overview of the biological roles of the included ncRNAs. In addition, we performed cell type-specific functional enrichment analyses. However, owing to the lack of sufficient molecules, the enrichment analysis in PAFs could not be conducted. GO analysis and KEGG pathway enrichment in PASMCs and PAECs revealed several PH-associated terms, such as positive regulation of smooth muscle cell proliferation, positive regulation of endothelial cell proliferation, HIF-1 signaling pathway, and MAPK signaling pathway. The top 10 enriched GO biological process terms and KEGG pathways are shown in Figure 6.
FIGURE 6.
Functional analyses of the ncRNAs. (A,B). The top 10 enriched GO biological process terms and KEGG pathways in all cell types. (C,D) The top 10 enriched GO biological process terms and KEGG pathways in PASMCs. (E,F) The top 10 enriched GO biological process terms and KEGG pathways in PAECs. Node sizes indicate the number of genes enriched in functional clusters. Node colors are related to q values. GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
3.4 Key Non-coding RNA Subnetworks
We built three networks according to the cell types. Here, we discuss several important subnetworks, along with their components and interactions, to improve understanding of the roles of ncRNAs in PH. Subnetworks with more than five nodes were regarded as key subnetworks.
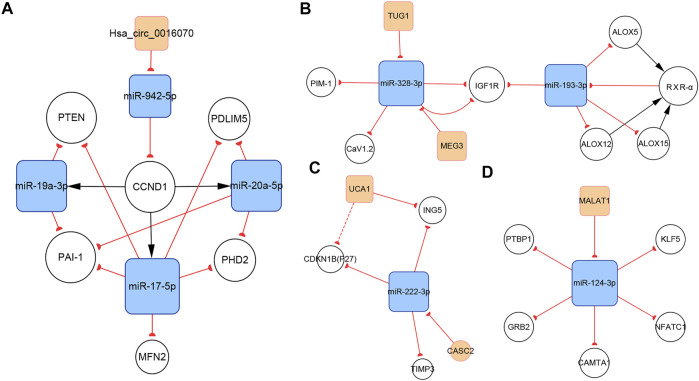
3.4.1 The Hsa_circ_0016070/miR-942-5p/CCND1 Subnetwork
CircRNAs are associated with various cardiovascular diseases. Hsa_circ_0016070 was the only circRNA included in our networks. This circRNA is located at chr1: 203595914-203702528, strand: +, promotes cell proliferation by mediating cell cycle progression, and is increased in PH patients (Zhou et al., 2019). CCND1 is an important regulator of the cell cycle. It interacts with cyclin-dependent kinase 4 (CDK4) to form the cyclin D1–CKD4 complex, which then inactivates retinoblastoma (Rb) protein and induces G0 progression to S phase (Matsushime et al., 1991). The subnetwork showed that hsa_circ_0016070 overexpression induced CCND1 expression by buffering miR-942-5p (Zhou et al., 2019). In addition, according to our network, CCND1 could induce the expression of miR-17-5p, miR-19a-3p, and miR-20a-5p, subsequently regulating the biological activities of PASMCs (Figure 7A). Given the considerable number of identified circRNAs, there should be other PH-related circRNAs. A microarray expression profile in thromboembolic pulmonary hypertension patients indicated that hsa_circ_0002062 and hsa_circ_0022342 might be the key circRNAs for the development of chronic thromboembolic pulmonary hypertension (Miao et al., 2017). However, this finding has not been verified by more reliable experimental methods.
FIGURE 7.
Several key ncRNA subnetworks. (A) The hsa_circ_0016070/miR-942-5p/CCND1 subnetwork. (B) The TUG1/MEG3/miR-328-3p/miR-193-3p subnetwork. (C) The CASC2/UCA1/miR-222-3p subnetwork. (D) The MALAT1/miR-124-3p subnetwork.
3.4.2 The TUG1/MEG3/miR-328-3p/miR-193-3p Subnetwork
Regulatory relationships are indicated by the connection lines in the subnetwork. According to this subnetwork (Figure 7B), both TUG1 and MEG3 can function as competing endogenous RNAs (ceRNAs) that sequester miR-328-3p. In the original studies, the TUG1/miR-328-3p and MEG3/miR-328-3p axes were identified (Wang D et al., 2019; Xing X.-Q et al., 2019). IGF1 is reported to inhibit PASMCs apoptosis and activate elastin in PASMCs. Thus, upregulating IGF1R via the TUG1/miR-328-3p and MEG3/miR-328-3p axes can induce PH by amplifying the pathogenic role of IGF1 (Wang S et al., 2019; Xing Y et al., 2019). Calcium voltage-gated channel subunit alpha1 C (CaV1.2), which contributes to vasoconstriction, is also a target gene of miR-328-3p in PASMCs (Guo et al., 2012), indicating that the TUG1/miR-328-3p and MEG3/miR-328-3p axes are involved in regulating pulmonary artery contraction and dilation. In addition, miR-328-3p can inhibit PASMC proliferation by targeting PIM-1 (Qian et al., 2016). Available data show that miR-193-3p has a shared target gene, IGF1R, with miR-328-3p, but no strong regulatory connection with miR-328-3p or TUG1 or MEG3. Thus, downregulation of miR-193-3p contributes to IGF1R overexpression as well. In addition, miR-193-3p is capable of negatively regulating multiple lipoxygenases, including ALOX5, ALOX12, and ALOX15. These lipoxygenases cause abnormal lipid metabolism, which not only directly accelerates the development of PH, but also induces the increasement of RXR-α. Moreover, miR-193-3p can be downregulated by RXR-α, which directly binds to the miR-193 promoter. Therefore, a feedback loop, which dramatically enhances abnormal miR-193-3p expression forms (Sharma et al., 2014).
3.4.3 The CASC2/UCA1/miR-222-3p Subnetwork
LncRNA CASC2 is downregulated in hypoxia-induced PASMCs. As a ceRNA of miR-222-3p, CASC2 reduces the expression of ING5, which is a target gene of miR-222-3p, ultimately promoting PASMC proliferation and migration (Han et al., 2020). P27 and TIMP3 are two additional target genes of miR-222-3p (Xu et al., 2017). P27, a member of the Cip/Kip family of cyclin-dependent kinase inhibitors, negatively regulates cell proliferation (Toyoshima and Hunter, 1994). Meanwhile, TIMP3 is a member of the TIMP family, which regulates cell proliferation, apoptosis, and migration via both MMP-dependent or MMP-independent pathways (Zhou et al., 2015). The present subnetwork links CACS2 to P27 and TIMP3 via miR-222-3p, further elaborating the mechanisms of PH (Figure 7C).
UCA1 is the other lncRNA in this subnetwork and is highly expressed in hypoxia-induced PASMCs. Studies indicate that UCA1 does not interact with miR-222-3p, but directly inhibits ING5 by competing with ING5 mRNA for hnRNP I, which binds to ING5 mRNA and enhances its translation. Thus, UCA1 overexpression results in the downregulation of ING5 mRNA expression (Zhu T.-T. et al., 2019). The same regulatory pattern has been found between UCA1 and P27 in breast tumor studies (Huang et al., 2014). This interaction may also work in PH and partly contributes to P27 downregulation (Figure 7C).
3.4.4 The MALAT1/miR-124-3p Subnetwork
LncRNA MALAT1, located at 11q13, is an 8.5-kb molecule that was identified by Ji et al. in a cancer study (Ji et al., 2003). Emerging evidence indicates that MALAT1 plays important roles in various diseases, including PH. Wang et al. reported that MALAT1 is highly expressed in pulmonary artery tissues and PASMCs from patients with PH. MALAT1 controls PASMC proliferation and migration by binding to miR-124-3p, which directly targets KLF5 (Wang D et al., 2019). Kang et al. showed that miR-124-3p also targets three regulators of the NFAT pathway, including NFATc1, CAMTA1, and PTBP1 (Kang B.-Y et al., 2013). The downregulation of miR-124-3p induces PASMC proliferation and reverses the differentiated PASMC phenotype by activating the NFAT pathway. In addition to its role in PASMCs, miR-124-3p also regulates the biological behaviors of PAH endothelial cells (PAH ECs) and PAFs. Studies have confirmed the role of the miR-124-3p/PTBP1 axis in PAH ECs and PAFs (Caruso et al., 2017; Wang et al., 2014; Zhang H et al., 2017). Downregulating miR-124-3p activates PTBP1 expression, which promotes aerobic glycolysis by increasing the PKM2/PKM1 ratio, subsequently inducing PAH EC and PAF proliferation (Anastasiou et al., 2012). Li et al. reported another target of miR-124-3p, GRB2, which enhanced the proliferation of multiple human cells (Li L et al., 2017; Figure 7D).
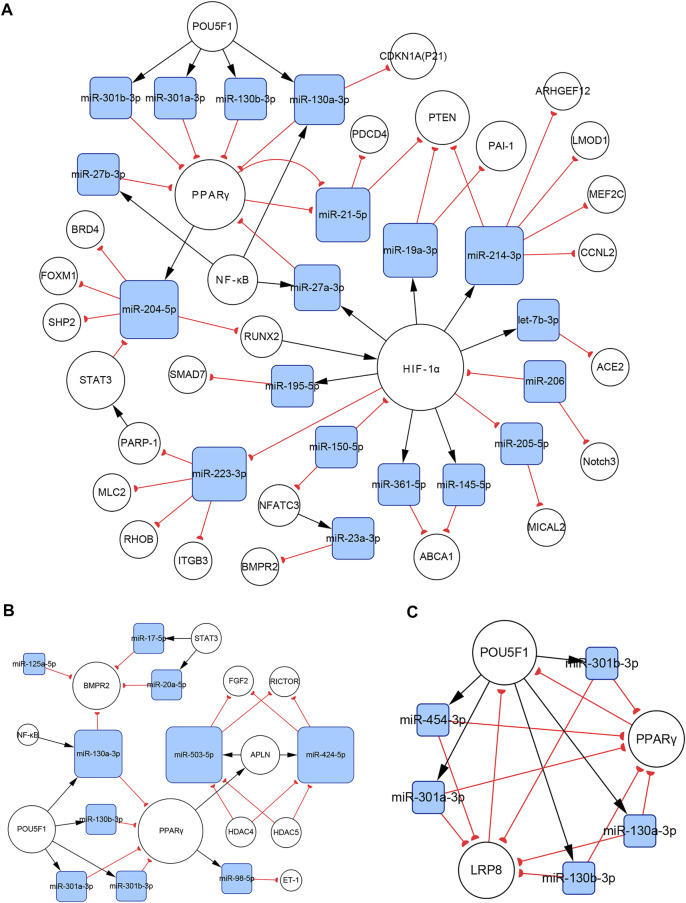
3.4.5 Subnetworks of the miR-130/301 Family
There are complicated relationships between the miR-130/301 family and other functional molecules associated with the pathogenesis of PH. In the present study, we found that subnetworks of the miR-130/301 family were involved in multiple biological behaviors, such as proliferation, apoptosis, and migration in PASMCs, PAECs, and PAFs. In addition, these subnetworks also mediated the crosstalk of these pulmonary artery cells.
In PASMCs, the miR-130/301 family is involved in many regulatory axes. Among them, the POU5F1/miR-130/301 family/PPARγ axis, which regulates the expression of miR-204-5p and miR-21-5p, is the most explicitly elaborated axis. According to our studies, the identified target genes of the two miRNAs in PASMCs include BRD4, FOXM1, PSCD4, PTEN, RUNX2, and SHP2, which control cell proliferation, apoptosis, differentiation, and mitochondrial function (Courboulin et al., 2011; Meloche et al., 2015a; Green et al., 2015, 2017; Ruffenach et al., 2016; Liu et al., 2017; Bourgeois et al., 2018a). In addition to the miR-130/301 family, miR-27a/b-3p, which is regulated by NF-κB (Xie et al., 2017), can also act as an upstream controller of PPARγ in PASMCs. Interestingly, the subnetwork analysis indicates that the miR-130/301 family indirectly promotes HIF-1α expression by sustaining the RUNX2 level (Ruffenach et al., 2016). Conversely, HIF-1α induces the expression of miR-27a-3p, which depresses the level of PPARγ (Camps et al., 2014). Thus, a feedback loop with PPARγ and HIF-1α forms. This loop leads to a persistent pathological status. Moreover, as a crucial pathogenic molecule for PH, HIF-1α can function through several miRNAs, including miR-145-5p, miR-19a-3p, miR-195-5p, miR-210-3p, miR-223-3p, and miR-361-5p, to regulate the expression of downstream proteins, eventually causing abnormal cellular behaviors (Agrawal et al., 2014; Gou et al., 2012; Meloche et al., 2015b; Zeng et al., 2018; Zhang X et al., 2018, Zhang H et al., 2019; Zhao et al., 2019; Figure 8A).
FIGURE 8.
The miR-130/301 family subnetworks in (A) PASMCs, (B) PAECs, and (C) PAFs. This miRNA family was involved in multiple processes, such as cell proliferation, apoptosis, migration, endothelial contraction, and matrix remodeling.
In PAECs, the miR-130/301 family also plays an important role. The POU5F1/miR-130/301 family/PPARγ axis indirectly regulates the expression of ET-1 and FGF2 via miR-98-5p and miR-424/503-5p, respectively (Kim et al., 2013; Zhang Y et al., 2018). The roles of ET-1 and FGF2 in PH are well established. ET-1 is synthesized primarily in endothelial cells and mediates pulmonary artery cell proliferation, migration, and constriction through two distinct G protein-coupled receptors: ETA and ETB (Clozel, 2016). Previous studies suggest that excessive FGF2 expression promotes PAEC proliferation by activating ERK1/2 and inhibits apoptosis by inducing BCL2 and BCL-xL activity (Tu et al., 2011). Furthermore, miR-130a-3p controls the level of BMPR2, which triggers idiopathic pulmonary artery hypertension (IPAH) and is involved in the development of other types of PH (Li Q et al., 2017). Considering that miRNAs from the same family have a homologous seed region sequence, other members from the miR-130/301 family may also regulate BMPR2 expression. The transcription of miR-130a-3p is controlled by NF-κB in PAECs. Thus, NF-κB and BMPR2 are linked by miR-130a-3p. In addition, miR-17a-5p, miR-20a-5p, and miR-125a-5p also mediate BMPR2 expression. Besides, two members from the miR-17-92 family, miR-17a-5p and miR-20a-5p, link STAT3 to BMPR2 (Brock et al., 2009; Huber et al., 2015; Figure 8B).
In PAFs, activation of the miR-130/301 family can induce cell proliferation and extracellular matrix remodeling by inhibiting PPARγ and LRP8. Meanwhile, matrix remodeling can activate POU5F1, which subsequently promotes miR-130/301 family expression (Bertero et al., 2015). Thus, a positive feedback circuit is activated that dramatically accelerates the development of PH (Figure 8C).
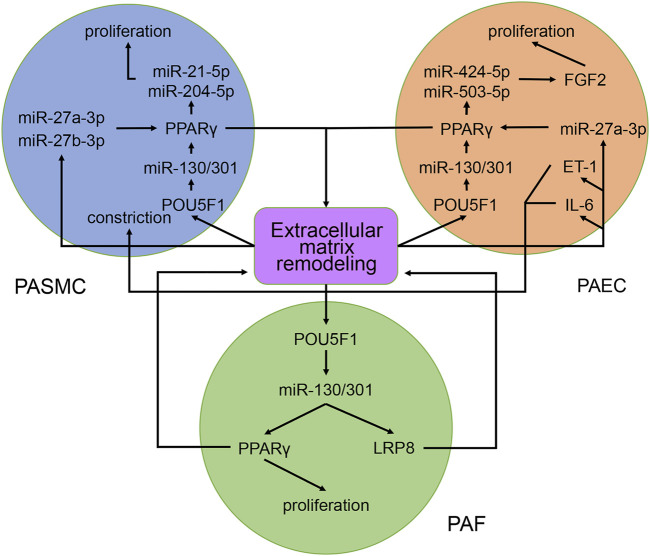
The roles of the miR-130/301 family in different pulmonary artery cell types are not independent. Rather, the miR-130/301 family contributes to crosstalk between these cells. Extracellular matrix remodeling, which can be induced by overexpression of the miR-130/301 family, promotes proliferation and contraction of pulmonary artery cells viamiR-130/301 family-dependent and -independent pathways. The remodeled extracellular matrix can activate the POU5F1/miR-130/301 family/PPARγ axis in PASMCs, PAECs, and PAFs, subsequently regulating downstream molecules such as miR-204-5p, miR-424-5p, miR-503-5p, and FGF2 (Bertero et al., 2015). Upregulating miR-424-5p and miR-503-5p or inhibiting FGF2 in PAECs can repress PASMC and PAF proliferation induced by conditioned media from PAECs, indicating that these molecules are involved in the crosstalk among different pulmonary vascular cells. The remodeled extracellular matrix can also induce the expression of the proliferative miRNA, miR-27a/b-3p, in PACEs, and PASMCs, as well as the expression of the vasoconstrictor ET-1, and the inflammatory cytokine IL-6 in PACEs (Bertero et al., 2014; Bertero et al., 2015; Figure 9).
FIGURE 9.
Contribution of the miR-130/301 family to the crosstalk between various pulmonary artery cells. The remodeled extracellular matrix induces proliferation and contraction in pulmonary artery cells via miR-130/301 family-dependent and -independent pathways. Meanwhile, matrix remodeling can be induced by overexpression of the miR-130/301 family.
4 Environmental Factors in Pulmonary Hypertension
Noncoding RNA interference is an important epigenetic mechanism. Recent evidence has identified the roles of epigenetic changes in the development of PH. These changes link the pathogenic genes of PH and environmental factors such as hypoxia, virus infection, and air pollution (Gamen et al., 2016). For example, BMPR2 is a transmembrane serine/threonine kinase receptor, which is essential for vascular homeostasis. Although mutations in the BMPR2 gene account for a considerable portion of patients with familial pulmonary artery hypertension (FPAH), only 20–30% of carriers with mutations in this gene suffer from PH, indicating that other factors contribute to the onset of the disease (Orriols et al., 2017; Zhao et al., 2019). According to our network, hypoxia can induce the expression of several miRNAs by HIF-1α, such as, miR-145-5p, miR-19a-3p, miR-191-5p, miR-214-3p, and miR-27a-3p (Agrawal et al., 2014; Camps et al., 2014; el Azzouzi et al., 2013; Song et al., 2014; Zhao et al., 2019). Among them, miR-191-5p can increase cell proliferation, impair apoptosis, and induce phenotypic alteration through inhibiting BMPR2 expression, subsequently contributing to vascular remodeling (Song et al., 2014). Therefore, the HIF-1α/miR-191-5p/BMPR2 axis reveals the connection between hypoxia and BMPR2 expression and partially explains the incomplete penetrance of BMPR2 mutations in FPAH.
5 Potential Applications of Non-coding RNAs
Ultimately, studies on molecular mechanisms aim to inform clinical practices. NcRNAs are potential diagnostic biomarkers for PH. For example, circRNAs are not easily degraded, making them ideal serum biomarkers. Zhang et al. reported hsa_circ_0068481 overexpression in the serum from patients with IPAH. Furthermore, hsa_circ_0068481 expression is significantly correlated with 6-min walk distance, N-terminal pro-B-type natriuretic peptide, H2S, pulmonary hypertension risk stratification, right heart failure, and survival rate (Zhang et al., 2019a). However, because of the absence of an associated molecular mechanism, this circRNA was not included in our networks. NcRNAs may also act as potential therapeutic targets for PH. For example, Rothman et al. identified downregulation of miR-140-5p in a rat PH model. In vitro, miR-140-5p mimics suppressed PASMC proliferation and migration. In vivo, miR-140-5p mimics prevented the progression of established PH in rats (Rothman et al., 2016). The results are encouraging. However, ncRNA therapy is far from being applied in clinical settings, since a ncRNA may have diverse biofunctions. This means that when used as therapeutic agent, a ncRNA may cause adverse effects, some of which may even be life-threatening. In our opinion, carefully selected ncRNA targets and well-designed action sites can be helpful to avoid such adverse effects. These measures require a comprehensive and in-depth understanding of the mechanisms of ncRNAs in diseases. In this study, we constructed networks to demonstrate the current findings on ncRNAs from studies performed in PH patients and animal models. However, shortcomings of these studies, including the paucity of human data, sex bias, and heterogeneity of animal models, limit the translation of these findings into applications for human disease. Therefore, further studies should be performed to confirm these findings in different animal models and patient cohorts of PH. Additionally, large, well-designed, and unbiased clinical studies are required to illuminate further application of ncRNAs.
6 Conclusion
The roles of ncRNAs in PH remained unclear. In this study, we performed an extensive literature search and adopted uniform and strict criteria for the selection of each article to avoid biased outcomes. The ncRNA networks were constructed by assembling ncRNAs and their interacting RNAs or genes from included articles. These networks provide a better understanding of the roles of ncRNAs in PH and can be helpful in elucidating the potential clinical applications of ncRNAs.
Author Contributions
Conceptualization, HZ; writing—original draft preparation HZ and QZ; writing—review and editing, HZ and XL. All authors have read and agreed to the published version of the article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Agrawal R., Pandey P., Jha P., Dwivedi V., Sarkar C., Kulshreshtha R. (2014). Hypoxic Signature of Micrornas in Glioblastoma: Insights from Small Rna Deep Sequencing. BMC Genomics 15, 686. 10.1186/1471-2164-15-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D., Yu Y., Israelsen W. J., Jiang J.-K., Boxer M. B., Hong B. S., et al. (2012). Pyruvate Kinase M2 Activators Promote Tetramer Formation and Suppress Tumorigenesis. Nat. Chem. Biol. 8, 839–847. 10.1038/nchembio.1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicheva A., Ayon R. J., Zhao T., Ek Vitorin J. F., Pohl N. M., Yamamura A., et al. (2020). MicroRNA-mediated Downregulation of K+ Channels in Pulmonary Arterial Hypertension. Am. J. Physiology-Lung Cell Mol. Physiol. 318, L10–L26. 10.1152/ajplung.00010.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero T., Cottrill K. A., Lu Y., Haeger C. M., Dieffenbach P., Annis S., et al. (2015). Matrix Remodeling Promotes Pulmonary Hypertension through Feedback Mechanoactivation of the Yap/taz-Mir-130/301 Circuit. Cel Rep. 13, 1016–1032. 10.1016/j.celrep.2015.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero T., Lu Y., Annis S., Hale A., Bhat B., Saggar R., et al. (2014). Systems-level Regulation of Microrna Networks by Mir-130/301 Promotes Pulmonary Hypertension. J. Clin. Invest. 124, 3514–3528. 10.1172/jci74773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi R., Bao C., Jiang L., Liu H., Yang Y., Mei J., et al. (2015). MicroRNA-27b Plays a Role in Pulmonary Arterial Hypertension by Modulating Peroxisome Proliferator-Activated Receptor γ Dependent Hsp90-eNOS Signaling and Nitric Oxide Production. Biochem. Biophysical Res. Commun. 460, 469–475. 10.1016/j.bbrc.2015.03.057 [DOI] [PubMed] [Google Scholar]
- Botti G., Marra L., Malzone M., Anniciello A., Botti C., Franco R., et al. (2016). Lncrna Hotair as Prognostic Circulating Marker and Potential Therapeutic Target in Patients with Tumor Diseases. Cdt 18, 27–34. 10.2174/1389450117666151209122950 [DOI] [PubMed] [Google Scholar]
- Bourgeois A., Lambert C., Habbout K., Ranchoux B., Paquet-Marceau S., Trinh I., et al. (2018a). Foxm1 Promotes Pulmonary Artery Smooth Muscle Cell Expansion in Pulmonary Arterial Hypertension. J. Mol. Med. 96, 223–235. 10.1007/s00109-017-1619-0 [DOI] [PubMed] [Google Scholar]
- Bourgeois A., Omura J., Habbout K., Bonnet S., Boucherat O. (2018b). Pulmonary Arterial Hypertension: New Pathophysiological Insights and Emerging Therapeutic Targets. Int. J. Biochem. Cel Biol. 104, 9–13. 10.1016/j.biocel.2018.08.015 [DOI] [PubMed] [Google Scholar]
- Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., et al. (2008). A Double-Negative Feedback Loop between Zeb1-Sip1 and the Microrna-200 Family Regulates Epithelial-Mesenchymal Transition. Cancer Res. 68, 7846–7854. 10.1158/0008-5472.Can-08-1942 [DOI] [PubMed] [Google Scholar]
- Brock M., Haider T. J., Vogel J., Gassmann M., Speich R., Trenkmann M., et al. (2015). The Hypoxia-Induced Microrna-130a Controls Pulmonary Smooth Muscle Cell Proliferation by Directly Targeting Cdkn1a. Int. J. Biochem. Cel Biol. 61, 129–137. 10.1016/j.biocel.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Brock M., Trenkmann M., Gay R. E., Michel B. A., Gay S., Fischler M., et al. (2009). Interleukin-6 Modulates the Expression of the Bone Morphogenic Protein Receptor Type Ii through a Novel Stat3-Microrna Cluster 17/92 Pathway. Circ. Res. 104, 1184–1191. 10.1161/circresaha.109.197491 [DOI] [PubMed] [Google Scholar]
- Cai Z., Li J., Zhuang Q., Zhang X., Yuan A., Shen L., et al. (2018). MiR-125a-5p Ameliorates Monocrotaline-Induced Pulmonary Arterial Hypertension by Targeting the TGF-Β1 and IL-6/STAT3 Signaling Pathways. Exp. Mol. Med. 50, 1–11. 10.1038/s12276-018-0068-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps C., Saini H. K., Mole D. R., Choudhry H., Reczko M., Guerra-Assunção J., et al. (2014). Integrated Analysis of Microrna and Mrna Expression and Association with Hif Binding Reveals the Complexity of Microrna Expression Regulation under Hypoxia. Mol. Cancer 13, 28. 10.1186/1476-4598-13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso P., Dunmore B. J., Schlosser K., Schoors S., Dos Santos C., Perez-Iratxeta C., et al. (2017). Identification of Microrna-124 as a Major Regulator of Enhanced Endothelial Cell Glycolysis in Pulmonary Arterial Hypertension via Ptbp1 (Polypyrimidine Tract Binding Protein) and Pyruvate Kinase M2. Circulation 136, 2451–2467. 10.1161/circulationaha.117.028034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.-C., Yu D., Lee Y.-S., Wentzel E. A., Arking D. E., West K. M., et al. (2008). Widespread Microrna Repression by Myc Contributes to Tumorigenesis. Nat. Genet. 40, 43–50. 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelladurai P., Seeger W., Pullamsetti S. S. (2016). Epigenetic Mechanisms in Pulmonary Arterial Hypertension: The Need for Global Perspectives. Eur. Respir. Rev. 25, 135–140. 10.1183/16000617.0036-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Cui X., Li L., Qu J., Raj J. U., Gou D. (2017). Mir-339 Inhibits Proliferation of Pulmonary Artery Smooth Muscle Cell by Targeting Fgf Signaling. Physiol. Rep. 5, e13441. 10.14814/phy2.13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.-H., Dasgupta A., Lin J., Potus F., Bonnet S., Iremonger J., et al. (2018). Epigenetic Dysregulation of the Dynamin-Related Protein 1 Binding Partners MiD49 and MiD51 Increases Mitotic Mitochondrial Fission and Promotes Pulmonary Arterial Hypertension. Circulation 138, 287–304. 10.1161/circulationaha.117.031258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Shen C., Zhang Y., Shu H. (2017). MicroRNA-150 Attenuates Hypoxia-Induced Excessive Proliferation and Migration of Pulmonary Arterial Smooth Muscle Cells through Reducing HIF-1α Expression. Biomed. Pharmacother. 93, 861–868. 10.1016/j.biopha.2017.07.028 [DOI] [PubMed] [Google Scholar]
- Chen S., Yu C., Lu R., Song T., Wang X., Tang W., et al. (2019). Mir-107 Inhibits Pdgf-Bb-Induced Proliferation of Human Pulmonary Arterial Smooth Muscle Cells and Migration through Targeting Nor1. Int. J. Clin. Exp. Pathol. 12, 1599–1608. [PMC free article] [PubMed] [Google Scholar]
- Chen T., Huang J. B., Dai J., Zhou Q., Raj J. U., Zhou G. (2018). Pai-1 Is a Novel Component of the Mir-17∼92 Signaling that Regulates Pulmonary Artery Smooth Muscle Cell Phenotypes. Am. J. Physiology-Lung Cell Mol. Physiol. 315, L149–L161. 10.1152/ajplung.00137.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Zhou G., Zhou Q., Tang H., Ibe J. C. F., Cheng H., et al. (2015). Loss of MicroRNA-17∼92 in Smooth Muscle Cells Attenuates Experimental Pulmonary Hypertension via Induction of PDZ and LIM Domain 5. Am. J. Respir. Crit. Care Med. 191, 678–692. 10.1164/rccm.201405-0941OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Zhou Q., Tang H., Bozkanat M., Yuan J. X. J., Raj J. U., et al. (2016). miR‐17/20 Controls Prolyl Hydroxylase 2 (PHD2)/Hypoxia‐Inducible Factor 1 (HIF1) to Regulate Pulmonary Artery Smooth Muscle Cell Proliferation. Jaha 5, e004510. 10.1161/jaha.116.004510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G., He L., Zhang Y. (2020). Lincrna-cox2 Promotes Pulmonary Arterial Hypertension by Regulating the Let-7a-Mediated Stat3 Signaling Pathway. Mol. Cel Biochem 475, 239–247. 10.1007/s11010-020-03877-6 [DOI] [PubMed] [Google Scholar]
- Clozel M. (2016). Endothelin Research and the Discovery of Macitentan for the Treatment of Pulmonary Arterial Hypertension. Am. J. Physiology-Regulatory, Integr. Comp. Physiol. 311, R721–R726. 10.1152/ajpregu.00475.2015 [DOI] [PubMed] [Google Scholar]
- Correia de Sousa M., Gjorgjieva M., Dolicka D., Sobolewski C., Foti M. (2019). Deciphering Mirnas' Action through Mirna Editing. Ijms 20, 6249. 10.3390/ijms20246249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courboulin A., Paulin R., Giguère N. J., Saksouk N., Perreault T., Meloche J., et al. (2011). Role for Mir-204 in Human Pulmonary Arterial Hypertension. J. Exp. Med. 208, 535–548. 10.1084/jem.20101812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. N., Hilyard A. C., Nguyen P. H., Lagna G., Hata A. (2010). Smad Proteins Bind a Conserved Rna Sequence to Promote Microrna Maturation by Drosha. Mol. Cel 39, 373–384. 10.1016/j.molcel.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B., Du J., Hu R., Wang A.-P., Wu W.-H., Hu C.-P., et al. (2016). MicroRNA-103/107 Is Involved in Hypoxia-Induced Proliferation of Pulmonary Arterial Smooth Muscle Cells by Targeting HIF-1β. Life Sci. 147, 117–124. 10.1016/j.lfs.2016.01.043 [DOI] [PubMed] [Google Scholar]
- Deng L., Blanco F. J., Stevens H., Lu R., Caudrillier A., McBride M., et al. (2015). Microrna-143 Activation Regulates Smooth Muscle and Endothelial Cell Crosstalk in Pulmonary Arterial Hypertension. Circ. Res. 117, 870–883. 10.1161/circresaha.115.306806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di X., Jin X., Li R., Zhao M., Wang K. (2019). Circrnas and Lung Cancer: Biomarkers and Master Regulators. Life Sci. 220, 177–185. 10.1016/j.lfs.2019.01.055 [DOI] [PubMed] [Google Scholar]
- el Azzouzi H., Leptidis S., Dirkx E., Hoeks J., van Bree B., Brand K., et al. (2013). The Hypoxia-Inducible MicroRNA Cluster miR-199a∼214 Targets Myocardial PPARδ and Impairs Mitochondrial Fatty Acid Oxidation. Cel Metab. 18, 341–354. 10.1016/j.cmet.2013.08.009 [DOI] [PubMed] [Google Scholar]
- Fu J., Bai P., Chen Y., Yu T., Li F. (2019). Inhibition of Mir-495 Improves Both Vascular Remodeling and Angiogenesis in Pulmonary Hypertension. J. Vasc. Res. 56, 97–106. 10.1159/000500024 [DOI] [PubMed] [Google Scholar]
- Galiè N., Humbert M., Vachiery J.-L., Gibbs S., Lang I., Torbicki A., et al. (2016). 2015 ESC/ERS Guidelines for the Diagnosis and Treatment of Pulmonary Hypertension. Eur. Heart J. 37, 67–119. 10.1093/eurheartj/ehv317 [DOI] [PubMed] [Google Scholar]
- Gamen E., Seeger W., Pullamsetti S. S. (2016). The Emerging Role of Epigenetics in Pulmonary Hypertension. Eur. Respir. J. 48, 903–917. 10.1183/13993003.01714-2015 [DOI] [PubMed] [Google Scholar]
- Gandellini P., Giannoni E., Casamichele A., Taddei M. L., Callari M., Piovan C., et al. (2014). Mir-205 Hinders the Malignant Interplay between Prostate Cancer Cells and Associated Fibroblasts. Antioxid. Redox Signaling 20, 1045–1059. 10.1089/ars.2013.5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou D., Ramchandran R., Peng X., Yao L., Kang K., Sarkar J., et al. (2012). Mir-210 Has an Antiapoptotic Effect in Pulmonary Artery Smooth Muscle Cells during Hypoxia. Am. J. Physiology-Lung Cell Mol. Physiol. 303, L682–L691. 10.1152/ajplung.00344.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D. E., Murphy T. C., Kang B.-Y., Bedi B., Yuan Z., Sadikot R. T., et al. (2017). Peroxisome Proliferator-Activated Receptor-γ Enhances Human Pulmonary Artery Smooth Muscle Cell Apoptosis through microRNA-21 and Programmed Cell Death 4. Am. J. Physiology-Lung Cell Mol. Physiol. 313, L371–L383. 10.1152/ajplung.00532.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D. E., Murphy T. C., Kang B.-Y., Searles C. D., Hart C. M. (2015). PPARγ Ligands Attenuate Hypoxia-Induced Proliferation in Human Pulmonary Artery Smooth Muscle Cells through Modulation of MicroRNA-21. PLoS One 10, e0133391. 10.1371/journal.pone.0133391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Qiu Z., Wei L., Yu X., Gao X., Jiang S., et al. (2012). The MicroRNA-328 Regulates Hypoxic Pulmonary Hypertension by Targeting at Insulin Growth Factor 1 Receptor and L-type Calcium Channel-Α1c. Hypertension 59, 1006–1013. 10.1161/hypertensionaha.111.185413 [DOI] [PubMed] [Google Scholar]
- Guo L., Yang Y., Liu J., Wang L., Li J., Wang Y., et al. (2014). Differentially Expressed Plasma Micrornas and the Potential Regulatory Function of Let-7b in Chronic Thromboembolic Pulmonary Hypertension. PLoS One 9, e101055. 10.1371/journal.pone.0101055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Liu Y., Yang C., Gao C., Guo X., Cheng J. (2020). Lncrna Casc2 Inhibits Hypoxia-Induced Pulmonary Artery Smooth Muscle Cell Proliferation and Migration by Regulating the Mir-222/ing5 axis. Cell Mol Biol Lett 25, 21. 10.1186/s11658-020-00215-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X., Ma C., Chen S., Dang J., Cheng X., Zhu D. (2018). Reverse the Down Regulation of Mir-92b-3p by Hypoxia Can Suppress the Proliferation of Pulmonary Artery Smooth Muscle Cells by Targeting Usp28. Biochem. Biophysical Res. Commun. 503, 3064–3077. 10.1016/j.bbrc.2018.08.095 [DOI] [PubMed] [Google Scholar]
- Hong Z., Chen K.-H., DasGupta A., Potus F., Dunham-Snary K., Bonnet S., et al. (2017). Microrna-138 and Microrna-25 Down-Regulate Mitochondrial Calcium Uniporter, Causing the Pulmonary Arterial Hypertension Cancer Phenotype. Am. J. Respir. Crit. Care Med. 195, 515–529. 10.1164/rccm.201604-0814OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou N., Watabe K., Lu Z., Wu F., Xu M., et al. (2014). Long Non-coding Rna uca1 Promotes Breast Tumor Growth by Suppression of P27 (Kip1). Cell Death Dis 5–e1008. e1008. 10.1038/cddis.2013.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber L. C., Ulrich S., Leuenberger C., Gassmann M., Vogel J., von Blotzheim L. G., et al. (2015). Featured Article: Microrna-125a in Pulmonary Hypertension: Regulator of a Proliferative Phenotype of Endothelial Cells. Exp. Biol. Med. (Maywood) 240, 1580–1589. 10.1177/1535370215579018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannone L., Zhao L., Dubois O., Duluc L., Rhodes C. J., Wharton J., et al. (2014). Mir-21/ddah1 Pathway Regulates Pulmonary Vascular Responses to Hypoxia. Biochem. J. 462, 103–112. 10.1042/bj20140486 [DOI] [PubMed] [Google Scholar]
- Inoue K., Fry E. A. (2015). Aberrant Expression of Cyclin D1 in Cancer. Signal. Transduction Insights 4, STI.S30306–13. 10.4137/sti.S30306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali S., Ramanathan G. K., Parthasarathy P. T., Aljubran S., Galam L., Yunus A., et al. (2012). Mir-206 Regulates Pulmonary Artery Smooth Muscle Cell Proliferation and Differentiation. PLoS One 7, e46808. 10.1371/journal.pone.0046808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandl K., Thekkekara Puthenparampil H., Marsh L. M., Hoffmann J., Wilhelm J., Veith C., et al. (2019). Long Non‐coding RNAs Influence the Transcriptome in Pulmonary Arterial Hypertension: the Role ofPAXIP1‐AS1. J. Pathol. 247, 357–370. 10.1002/path.5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P. M., et al. (2003). MALAT-1, a Novel Noncoding RNA, and Thymosin β4 Predict Metastasis and Survival in Early-Stage Non-small Cell Lung Cancer. Oncogene 22, 8031–8041. 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- Jiang J., Xia Y., Liang Y., Yang M., Zeng W., Zeng X. (2018). Mir-190a-5p Participates in the Regulation of Hypoxia-Induced Pulmonary Hypertension by Targeting Klf15 and Can Serve as a Biomarker of Diagnosis and Prognosis in Chronic Obstructive Pulmonary Disease Complicated with Pulmonary Hypertension. Copd Vol. 13, 3777–3790. 10.2147/copd.S182504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Pang T., Nelin L. D., Wang W., Wang Y., Yan J., et al. (2015). Mkp-1 Is a Target of Mir-210 and Mediate the Negative Regulation of Mir-210 Inhibitor on Hypoxic Hpasmc Proliferation. Cell Biol Int 39, 113–120. 10.1002/cbin.10339 [DOI] [PubMed] [Google Scholar]
- Kang B.-Y., Park K. K., Green D. E., Bijli K. M., Searles C. D., Sutliff R. L., et al. (2013). Hypoxia Mediates Mutual Repression between microRNA-27a and PPARγ in the Pulmonary Vasculature. PLoS One 8, e79503. 10.1371/journal.pone.0079503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B.-Y., Park K. K., Kleinhenz J. M., Murphy T. C., Green D. E., Bijli K. M., et al. (2016). Peroxisome Proliferator-Activated Receptor γ and microRNA 98 in Hypoxia-Induced Endothelin-1 Signaling. Am. J. Respir. Cel Mol Biol 54, 136–146. 10.1165/rcmb.2014-0337OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Peng X., Zhang X., Wang Y., Zhang L., Gao L., et al. (2013). Microrna-124 Suppresses the Transactivation of Nuclear Factor of Activated T Cells by Targeting Multiple Genes and Inhibits the Proliferation of Pulmonary Artery Smooth Muscle Cells. J. Biol. Chem. 288, 25414–25427. 10.1074/jbc.M113.460287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kang Y., Kojima Y., Lighthouse J. K., Hu X., Aldred M. A., et al. (2013). An Endothelial Apelin-Fgf Link Mediated by Mir-424 and Mir-503 Is Disrupted in Pulmonary Arterial Hypertension. Nat. Med. 19, 74–82. 10.1038/nm.3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kang H. (2019). Hypoxia Promotes Vascular Smooth Muscle Cell Proliferation through Microrna-Mediated Suppression of Cyclin-dependent Kinase Inhibitors. Cells 8, 802. 10.3390/cells8080802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S., Peng F., Li M.-L., Duan W.-B., Peng C.-Q., Wu S.-J. (2020). Lncrna-smilr Modulates Rhoa/rock Signaling by Targeting Mir-141 to Regulate Vascular Remodeling in Pulmonary Arterial Hypertension. Am. J. Physiology-Heart Circulatory Physiol. 319, H377–H391. 10.1152/ajpheart.00717 [DOI] [PubMed] [Google Scholar]
- Leisegang M. S., Fork C., Josipovic I., Richter F. M., Preussner J., Hu J., et al. (2017). Long Noncoding Rna Mantis Facilitates Endothelial Angiogenic Function. Circulation 136, 65–79. 10.1161/circulationaha.116.02699110.1152/ajpheart.00717.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kim I.-K., Chiasson V., Chatterjee P., Gupta S. (2017). NF-κB Mediated miR-130a Modulation in Lung Microvascular Cell Remodeling: Implication in Pulmonary Hypertension. Exp. Cel Res. 359, 235–242. 10.1016/j.yexcr.2017.07.024 [DOI] [PubMed] [Google Scholar]
- Li Q., Qian Z., Wang L. (2017). Pri-microrna-124 Rs531564 Polymorphism Minor Allele Increases the Risk of Pulmonary Artery Hypertension by Abnormally Enhancing Proliferation of Pulmonary Artery Smooth Muscle Cells. Copd 12, 1351–1361. 10.2147/copd.S99318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhou X., Zhou X. (2019). Downregulation of miR-98 C-ontributes to H-ypoxic P-ulmonary H-ypertension by T-argeting ALK1. Mol. Med. Rep. 20, 2167–2176. 10.3892/mmr.2019.10482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-S., Ran Y.-J., Zhang D.-D., Li S.-Z., Zhu D. (2014). MicroRNA-190 Regulates Hypoxic Pulmonary Vasoconstriction by Targeting a Voltage-Gated K+Channel in Arterial Smooth Muscle Cells. J. Cel. Biochem. 115, 1196–1205. 10.1002/jcb.24771 [DOI] [PubMed] [Google Scholar]
- Li S., Ran Y., Zhang D., Chen J., Li S., Zhu D. (2013). Microrna-138 Plays a Role in Hypoxic Pulmonary Vascular Remodelling by Targeting Mst1. Biochem. J. 452, 281–291. 10.1042/bj20120680 [DOI] [PubMed] [Google Scholar]
- Li Y., Li L., Qian Z., Lin B., Chen J., Luo Y., et al. (2018). Phosphatidylinositol 3‐Kinase-DNA Methyltransferase 1-miR‐1281-Histone Deacetylase 4 Regulatory Axis Mediates Platelet‐Derived Growth Factor-Induced Proliferation and Migration of Pulmonary Artery Smooth Muscle Cells. Jaha 7, e007572. 10.1161/jaha.117.007572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ren W., Wang X., Yu X., Cui L., Li X., et al. (2019). Microrna-150 Relieves Vascular Remodeling and Fibrosis in Hypoxia-Induced Pulmonary Hypertension. Biomed. Pharmacother. 109, 1740–1749. 10.1016/j.biopha.2018.11.058 [DOI] [PubMed] [Google Scholar]
- Lin Z., Murtaza I., Wang K., Jiao J., Gao J., Li P.-F. (2009). Mir-23a Functions Downstream of Nfatc3 to Regulate Cardiac Hypertrophy. Proc. Natl. Acad. Sci. 106, 12103–12108. 10.1073/pnas.0811371106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Liu Y., Li B., Yang M., Liu Y., Su J. (2019). Role of miR‐223‐3p in Pulmonary Arterial Hypertension via Targeting ITGB3 in the ECM Pathway. Cell Prolif 52, e12550. 10.1111/cpr.12550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Hao P., Xu J., Wang L., Wang Y., Han R., et al. (2018). Upregulation of Microrna-17-5p Contributes to Hypoxia-Induced Proliferation in Human Pulmonary Artery Smooth Muscle Cells through Modulation of P21 and Pten. Respir. Res. 19, 200. 10.1186/s12931-018-0902-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.-M., Jia Y., Zhang Y.-x., Yan J., Liao N., Li X.-h., et al. (2019). Dysregulation of Mir-135a-5p Promotes the Development of Rat Pulmonary Arterial Hypertension In Vivo and In Vitro . Acta Pharmacol. Sin 40, 477–485. 10.1038/s41401-018-0076-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Tao Y., Chen M., Yu J., Li W.-J., Tao L., et al. (2016). Upregulation of Microrna-214 Contributes to the Development of Vascular Remodeling in Hypoxia-Induced Pulmonary Hypertension via Targeting Ccnl2. Sci. Rep. 6, 24661. 10.1038/srep24661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yin T., Yan W., Si R., Wang B., Chen M., et al. (2017). Dysregulation of Microrna-214 and Pten Contributes to the Pathogenesis of Hypoxic Pulmonary Hypertension. Copd Vol. 12, 1781–1791. 10.2147/copd.S104627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. J., Zhang H., Xing F., Tang B., Wu S. L., Xuan L., et al. (2018). MicroRNA-138 P-romotes P-roliferation and S-uppresses M-itochondrial D-epolarization in H-uman P-ulmonary A-rtery S-mooth M-uscle C-ells through T-argeting TASK-1. Mol. Med. Rep. 17, 3021–3027. 10.3892/mmr.2017.8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-C., Chuang S.-M., Hsu C.-J., Tsai C.-H., Wang S.-W., Tang C.-H. (2014). Ctgf Increases Vascular Endothelial Growth Factor-dependent Angiogenesis in Human Synovial Fibroblasts by Increasing Mir-210 Expression. Cel Death Dis 5, e1485. 10.1038/cddis.2014.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Zou X.-Z., Huang N., Ge X.-Y., Yao M.-Z., Liu H., et al. (2019a). Down-regulation of Mir-204 Attenuates Endothelial-Mesenchymal Transition by Enhancing Autophagy in Hypoxia-Induced Pulmonary Hypertension. Eur. J. Pharmacol. 863, 172673. 10.1016/j.ejphar.2019.172673 [DOI] [PubMed] [Google Scholar]
- Liu T., Zou X.-Z., Huang N., Ge X.-Y., Yao M.-Z., Liu H., et al. (2019b). Mir-27a Promotes Endothelial-Mesenchymal Transition in Hypoxia-Induced Pulmonary Arterial Hypertension by Suppressing Bmp Signaling. Life Sci. 227, 64–73. 10.1016/j.lfs.2019.04.038 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu G., Zhang H., Wang J. (2016). Mirna-199a-5p Influences Pulmonary Artery Hypertension via Downregulating Smad3. Biochem. Biophysical Res. Commun. 473, 859–866. 10.1016/j.bbrc.2016.03.140 [DOI] [PubMed] [Google Scholar]
- Lu Z., Li S., Zhao S., Fa X. (2016). Upregulated Mir-17 Regulates Hypoxia-Mediated Human Pulmonary Artery Smooth Muscle Cell Proliferation and Apoptosis by Targeting Mitofusin 2. Med. Sci. Monit. 22, 3301–3308. 10.12659/msm.900487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Zhang C., Ma M., Zhang L., Zhang L., Zhang F., et al. (2017). Mir-125a Regulates Mitochondrial Homeostasis through Targeting Mitofusin 1 to Control Hypoxic Pulmonary Vascular Remodeling. J. Mol. Med. 95, 977–993. 10.1007/s00109-017-1541-5 [DOI] [PubMed] [Google Scholar]
- Matsushime H., Roussel M. F., Ashmun R. A., Sherr C. J. (1991). Colony-stimulating Factor 1 Regulates Novel Cyclins during the G1 Phase of the Cell Cycle. Cell 65, 701–713. 10.1016/0092-8674(91)90101-4 [DOI] [PubMed] [Google Scholar]
- Meloche J., Le Guen M., Potus F., Vinck J., Ranchoux B., Johnson I., et al. (2015b). Mir-223 Reverses Experimental Pulmonary Arterial Hypertension. Am. J. Physiology-Cell Physiol. 309, C363–C372. 10.1152/ajpcell.00149.2015 [DOI] [PubMed] [Google Scholar]
- Meloche J., Pflieger A., Vaillancourt M., Paulin R., Potus F., Zervopoulos S., et al. (2014). Role for DNA Damage Signaling in Pulmonary Arterial Hypertension. Circulation 129, 786–797. 10.1161/circulationaha.113.006167 [DOI] [PubMed] [Google Scholar]
- Meloche J., Potus F., Vaillancourt M., Bourgeois A., Johnson I., Deschamps L., et al. (2015a). Bromodomain-Containing Protein 4. Circ. Res. 117, 525–535. 10.1161/circresaha.115.307004 [DOI] [PubMed] [Google Scholar]
- Miao R., Wang Y., Wan J., Leng D., Gong J., Li J., et al. (2017). Microarray Expression Profile of Circular Rnas in Chronic Thromboembolic Pulmonary Hypertension. Medicine (Baltimore) 96, e7354. 10.1097/md.0000000000007354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X., Chen Y., Tan J., Dai Y., Mao W., Qin G., et al. (2019). Microrna-221-3p Promotes Pulmonary Artery Smooth Muscle Cells Proliferation by Targeting Axin2 during Pulmonary Arterial Hypertension. Vasc. Pharmacol. 116, 24–35. 10.1016/j.vph.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Orriols M., Gomez-Puerto M. C., Ten Dijke P. (2017). Bmp Type Ii Receptor as a Therapeutic Target in Pulmonary Arterial Hypertension. Cell. Mol. Life Sci. 74, 2979–2995. 10.1007/s00018-017-2510-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh V. N., Jin R. C., Rabello S., Gulbahce N., White K., Hale A., et al. (2012). MicroRNA-21 Integrates Pathogenic Signaling to Control Pulmonary Hypertension. Circulation 125, 1520–1532. 10.1161/circulationaha.111.060269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Li Y., Chen J., Li X., Gou D. (2017). Mir-4632 Mediates Pdgf-Bb-Induced Proliferation and Antiapoptosis of Human Pulmonary Artery Smooth Muscle Cells via Targeting Cjun. Am. J. Physiology-Cell Physiol. 313, C380–C391. 10.1152/ajpcell.00061.2017 [DOI] [PubMed] [Google Scholar]
- Qian Z., Li Y., Yang H., Chen J., Li X., Gou D. (2018). Pdgfbb Promotes Proliferation and Migration via Regulating Mir-1181/stat3 axis in Human Pulmonary Arterial Smooth Muscle Cells. Am. J. Physiology-Lung Cell Mol. Physiol. 315, L965–L976. 10.1152/ajplung.00224.2018 [DOI] [PubMed] [Google Scholar]
- Qian Z., Zhang L., Chen J., Li Y., Kang K., Qu J., et al. (2016). Mir-328 Targeting Pim-1 Inhibits Proliferation and Migration of Pulmonary Arterial Smooth Muscle Cells in Pdgfbb Signaling Pathway. Oncotarget 7, 54998–55011. 10.18632/oncotarget.10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokavec M., Öner M. G., Li H., Jackstadt R., Jiang L., Lodygin D., et al. (2014). Il-6r/stat3/mir-34a Feedback Loop Promotes Emt-Mediated Colorectal Cancer Invasion and Metastasis. J. Clin. Invest. 124, 1853–1867. 10.1172/jci73531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A. M. K., Arnold N. D., Pickworth J. A., Iremonger J., Ciuclan L., Allen R. M. H., et al. (2016). Microrna-140-5p and Smurf1 Regulate Pulmonary Arterial Hypertension. J. Clin. Invest. 126, 2495–2508. 10.1172/jci83361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffenach G., Chabot S., Tanguay V. F., Courboulin A., Boucherat O., Potus F., et al. (2016). Role for Runt-Related Transcription Factor 2 in Proliferative and Calcified Vascular Lesions in Pulmonary Arterial Hypertension. Am. J. Respir. Crit. Care Med. 194, 1273–1285. 10.1164/rccm.201512-2380OC [DOI] [PubMed] [Google Scholar]
- Sahoo S., Meijles D. N., Al Ghouleh I., Tandon M., Cifuentes-Pagano E., Sembrat J., et al. (2016). Mef2c-myocd and Leiomodin1 Suppression by Mirna-214 Promotes Smooth Muscle Cell Phenotype Switching in Pulmonary Arterial Hypertension. PLoS One 11, e0153780. 10.1371/journal.pone.0153780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang H.-y., Jin Y.-l., Zhang W.-q., Chen L.-b. (2016). Downregulation of Microrna-637 Increases Risk of Hypoxia-Induced Pulmonary Hypertension by Modulating Expression of Cyclin Dependent Kinase 6 (Cdk6) in Pulmonary Smooth Muscle Cells. Med. Sci. Monit. 22, 4066–4072. 10.12659/msm.897254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte J. H., Horn S., Otto T., Samans B., Heukamp L. C., Eilers U.-C., et al. (2008). Mycn Regulates Oncogenic Micrornas in Neuroblastoma. Int. J. Cancer 122, 699–704. 10.1002/ijc.23153 [DOI] [PubMed] [Google Scholar]
- Sharma S., Umar S., Potus F., Iorga A., Wong G., Meriwether D., et al. (2014). Apolipoprotein A-I Mimetic Peptide 4f Rescues Pulmonary Hypertension by Inducing Microrna-193-3p. Circulation 130, 776–785. 10.1161/circulationaha.114.007405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Ren H., Gao S., Zhao X., Zhang H., Hao J. (2014). The Clinical Significance and Regulation Mechanism of Hypoxia-Inducible Factor-1 and Mir-191 Expression in Pancreatic Cancer. Tumor Biol. 35, 11319–11328. 10.1007/s13277-014-2452-5 [DOI] [PubMed] [Google Scholar]
- Su H., Xu X., Yan C., Shi Y., Hu Y., Dong L., et al. (2018). LncRNA H19 Promotes the Proliferation of Pulmonary Artery Smooth Muscle Cells through AT1R via Sponging Let-7b in Monocrotaline-Induced Pulmonary Arterial Hypertension. Respir. Res. 19, 254. 10.1186/s12931-018-0956-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Lin P., Chen Y., Yu H., Ren S., Wang J., et al. (2020). Mir-182-3p/myadm Contribute to Pulmonary Artery Hypertension Vascular Remodeling via a Klf4/p21-dependent Mechanism. Theranostics 10, 5581–5599. 10.7150/thno.44687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysol J. R., Chen J., Singla S., Zhao S., Comhair S., Natarajan V., et al. (2018). Micro-rna-1 Is Decreased by Hypoxia and Contributes to the Development of Pulmonary Vascular Remodeling via Regulation of Sphingosine Kinase 1. Am. J. Physiology-Lung Cell Mol. Physiol. 314, L461–L472. 10.1152/ajplung.00057.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K., Yamakuchi M., Matsuyama T., Kondo K., Uchida A., Misono S., et al. (2018). Il-13 Enhances Mesenchymal Transition of Pulmonary Artery Endothelial Cells via Down-Regulation of Mir-424/503 In Vitro . Cell Signal. 42, 270–280. 10.1016/j.cellsig.2017.10.019 [DOI] [PubMed] [Google Scholar]
- Tan H., Yao H., Lie Z., Chen G., Lin S., Zhang Y. (2019). MicroRNA-30a-5p P-romotes P-roliferation and I-nhibits A-poptosis of H-uman P-ulmonary A-rtery E-ndothelial C-ells under H-ypoxia by T-argeting YKL-40. Mol. Med. Rep. 20, 236–244. 10.3892/mmr.2019.10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W., Sun W., Zhu H., Zhang J. (2019). Mir-205-5p Suppresses Pulmonary Vascular Smooth Muscle Cell Proliferation by Targeting Mical2-Mediated Erk1/2 Signaling. Microvasc. Res. 124, 43–50. 10.1016/j.mvr.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Toyoshima H., Hunter T. (1994). P27, a Novel Inhibitor of G1 Cyclin-Cdk Protein Kinase Activity, Is Related to P21. Cell 78, 67–74. 10.1016/0092-8674(94)90573-8 [DOI] [PubMed] [Google Scholar]
- Tu L., Dewachter L., Gore B., Fadel E., Dartevelle P., Simonneau G., et al. (2011). Autocrine Fibroblast Growth Factor-2 Signaling Contributes to Altered Endothelial Phenotype in Pulmonary Hypertension. Am. J. Respir. Cel Mol Biol 45, 311–322. 10.1165/rcmb.2010-0317OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith C., Schermuly R. T., Brandes R. P., Weissmann N. (2016). Molecular Mechanisms of Hypoxia-Inducible Factor-Induced Pulmonary Arterial Smooth Muscle Cell Alterations in Pulmonary Hypertension. J. Physiol. 594, 1167–1177. 10.1113/jp270689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M., Yokoyama S. (2014). Microrna-mediated Deadenylation in a Mammalian Cell-free System. Methods Mol. Biol. 1125, 341–351. 10.1007/978-1-62703-971-0_27 [DOI] [PubMed] [Google Scholar]
- Wallace E., Morrell N. W., Yang X. D., Long L., Stevens H., Nilsen M., et al. (2015). A Sex-specific Microrna-96/5-Hydroxytryptamine 1b axis Influences Development of Pulmonary Hypertension. Am. J. Respir. Crit. Care Med. 191, 1432–1442. 10.1164/rccm.201412-2148OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.-p., Li X.-h., Gong S.-x., Li W.-q., Hu C.-p., Zhang Z., et al. (2015). Mir-100 Suppresses Mtor Signaling in Hypoxia-Induced Pulmonary Hypertension in Rats. Eur. J. Pharmacol. 765, 565–573. 10.1016/j.ejphar.2015.09.031 [DOI] [PubMed] [Google Scholar]
- Wang D., Xu H., Wu B., Jiang S., Pan H., Wang R., et al. (2019). Long Non-coding RNA MALAT1 S-ponges miR-124-3p.1/KLF5 to P-romote P-ulmonary V-ascular R-emodeling and C-ell C-ycle P-rogression of P-ulmonary A-rtery H-ypertension. Int. J. Mol. Med. 44, 871–884. 10.3892/ijmm.2019.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang H., Li M., Frid M. G., Flockton A. R., McKeon B. A., et al. (2014). Microrna-124 Controls the Proliferative, Migratory, and Inflammatory Phenotype of Pulmonary Vascular Fibroblasts. Circ. Res. 114, 67–78. 10.1161/circresaha.114.301633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Qin R., Cheng Y. (2020). Lncrna-ang362 Promotes Pulmonary Arterial Hypertension by Regulating Mir-221 and Mir-222. Shock 53, 723–729. 10.1097/shk.0000000000001410 [DOI] [PubMed] [Google Scholar]
- Wang L. N., Yu W. C., Du C. H., Tong L., Cheng Z. Z. (2018). Hypoxia Is Involved in Hypoxic Pulmonary Hypertension through Inhibiting the Activation of Fgf2 by Mir-203. Eur. Rev. Med. Pharmacol. Sci. 22, 8866–8876. 10.26355/eurrev_201812_16655 [DOI] [PubMed] [Google Scholar]
- Wang P., Xu J., Hou Z., Wang F., Song Y., Wang J., et al. (2016). Mirna-34a Promotes Proliferation of Human Pulmonary Artery Smooth Muscle Cells by Targeting Pdgfra. Cell Prolif. 49, 484–493. 10.1111/cpr.12265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Ding X., Zhou S., Li M., Sun L., Xu X., et al. (2016). Microrna-26b Attenuates Monocrotaline-Induced Pulmonary Vascular Remodeling via Targeting Connective Tissue Growth Factor (Ctgf) and Cyclin D1 (Ccnd1). Oncotarget 7, 72746–72757. 10.18632/oncotarget.10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Cao W., Gao S., Nie X., Zheng X., Xing Y., et al. (2019). Tug1 Regulates Pulmonary Arterial Smooth Muscle Cell Proliferation in Pulmonary Arterial Hypertension. Can. J. Cardiol. 35, 1534–1545. 10.1016/j.cjca.2019.07.630 [DOI] [PubMed] [Google Scholar]
- Weber L., Rickli H., Joerg L., Weilenmann D., Brenner R., Taramasso M., et al. (2018). Haemodynamic Mechanisms and Long-Term Prognostic Impact of Pulmonary Hypertension in Patients with Severe Aortic Stenosis Undergoing Valve Replacement. Eur. J. Heart Fail. 21, 172–181. 10.1002/ejhf.1322 [DOI] [PubMed] [Google Scholar]
- White K., Lu Y., Annis S., Hale A. E., Chau B. N., Dahlman J. E., et al. (2015). Genetic and Hypoxic Alterations of the Micro RNA ‐210‐ ISCU 1/2 axis Promote Iron-Sulfur Deficiency and Pulmonary Hypertension. EMBO Mol. Med. 7, 695–713. 10.15252/emmm.201404511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Talbot C. C., Liu Q., Jing Z.-C., Damico R. L., Tuder R., et al. (2016). Identifying microRNAs Targeting Wnt/β-Catenin Pathway in End-Stage Idiopathic Pulmonary Arterial Hypertension. J. Mol. Med. 94, 875–885. 10.1007/s00109-016-1426-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Li S., Zhu Y., Liu L., Pan Y., Wang J., et al. (2017). MicroRNA-27a/b Mediates Endothelin-1-Induced PPARγ Reduction and Proliferation of Pulmonary Artery Smooth Muscle Cells. Cell Tissue Res 369, 527–539. 10.1007/s00441-017-2625-9 [DOI] [PubMed] [Google Scholar]
- Xing X.-Q., Li B., Xu S.-L., Liu J., Zhang C.-F., Yang J. (2019). Microrna-214-3p Regulates Hypoxia-Mediated Pulmonary Artery Smooth Muscle Cell Proliferation and Migration by Targeting Arhgef12. Med. Sci. Monit. 25, 5738–5746. 10.12659/msm.915709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zheng X., Fu Y., Qi J., Li M., Ma M., et al. (2019). Long Noncoding Rna-Maternally Expressed Gene 3 Contributes to Hypoxic Pulmonary Hypertension. Mol. Ther. 27, 2166–2181. 10.1016/j.ymthe.2019.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zheng X., Li G., Liao L., Cao W., Xing H., et al. (2015). MicroRNA-30c Contributes to the Development of Hypoxia Pulmonary Hypertension by Inhibiting Platelet-Derived Growth Factor Receptor β Expression. Int. J. Biochem. Cel Biol. 64, 155–166. 10.1016/j.biocel.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Xu G., Chen J., Jing G., Shalev A. (2013). Thioredoxin-interacting Protein Regulates Insulin Transcription through Microrna-204. Nat. Med. 19, 1141–1146. 10.1038/nm.3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Bei Y., Shen S., Zhang J., Lu Y., Xiao J., et al. (2017). Microrna-222 Promotes the Proliferation of Pulmonary Arterial Smooth Muscle Cells by Targeting P27 and Timp3. Cell Physiol Biochem 43, 282–292. 10.1159/000480371 [DOI] [PubMed] [Google Scholar]
- Yang Y. Z., Zhang Y. F., Yang L., Xu J., Mo X. M., Peng W. (2018). miR-760 M-ediates H-ypoxia-I-nduced P-roliferation and A-poptosis of H-uman P-ulmonary A-rtery S-mooth M-uscle C-ells via T-argeting TLR4. Int. J. Mol. Med. 42, 2437–2446. 10.3892/ijmm.2018.3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Xu M., Dong Y., Liu J., Li Y., Mao W., et al. (2018). 1,25(OH) 2 D 3 Attenuates Pulmonary Arterial Hypertension via microRNA-204 Mediated Tgfbr2/Smad Signaling. Exp. Cel Res. 362, 311–323. 10.1016/j.yexcr.2017.11.032 [DOI] [PubMed] [Google Scholar]
- Yu Z., Wang C., Wang M., Li Z., Casimiro M. C., Liu M., et al. (2008). A Cyclin D1/microrna 17/20 Regulatory Feedback Loop in Control of Breast Cancer Cell Proliferation. J. Cel Biol 182, 509–517. 10.1083/jcb.200801079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Xu M., Rong R., Mei Y., Cai W., Li L., et al. (2017). Mir-200c Regulates Endothelin-1 Induced Pasmcs Abnormal Proliferation and Apoptosis. IUBMB Life 69, 877–886. 10.1002/iub.1686 [DOI] [PubMed] [Google Scholar]
- Yue J., Guan J., Wang X., Zhang L., Yang Z., Ao Q., et al. (2013). MicroRNA-206 Is Involved in Hypoxia-Induced Pulmonary Hypertension through Targeting of the HIF-1α/Fhl-1 Pathway. Lab. Invest. 93, 748–759. 10.1038/labinvest.2013.63 [DOI] [PubMed] [Google Scholar]
- Yue Y., Zhang Z., Zhang L., Chen S., Guo Y., Hong Y. (2018). Mir-143 and Mir-145 Promote Hypoxia-Induced Proliferation and Migration of Pulmonary Arterial Smooth Muscle Cells through Regulating Abca1 Expression. Cardiovasc. Pathol. 37, 15–25. 10.1016/j.carpath.2018.08.003 [DOI] [PubMed] [Google Scholar]
- Zehendner C. M., Valasarajan C., Werner A., Boeckel J.-N., Bischoff F. C., John D., et al. (2020). Long Noncoding RNA TYKRIL Plays a Role in Pulmonary Hypertension via the P53-Mediated Regulation of PDGFRβ. Am. J. Respir. Crit. Care Med. 202, 1445–1457. 10.1164/rccm.201910-2041OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Zhang X., Kang K., Chen J., Wu Z., Huang J., et al. (2016). Microrna-223 Attenuates Hypoxia-Induced Vascular Remodeling by Targeting Rhob/mlc2 in Pulmonary Arterial Smooth Muscle Cells. Sci. Rep. 6, 24900. 10.1038/srep24900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z. H., Wu W. H., Peng Q., Sun Y. H., Liu J. X. (2019). MicroRNA-132 M-ediates P-roliferation and M-igration of P-ulmonary S-mooth M-uscle C-ells via T-argeting PTEN. Mol. Med. Rep. 19, 3823–3830. 10.3892/mmr.2019.10053 [DOI] [PubMed] [Google Scholar]
- Zeng Z., Yao J., Li Y., Xue Y., Zou Y., Shu Z., et al. (2018). Anti‐apoptosis Endothelial Cell‐secreted microRNA‐195‐5p Promotes Pulmonary Arterial Smooth Muscle Cell Proliferation and Migration in Pulmonary Arterial Hypertension. J. Cel. Biochem. 119, 2144–2155. 10.1002/jcb.26376 [DOI] [PubMed] [Google Scholar]
- Zhang C., Ma C., Zhang L., Zhang L., Zhang F., Ma M., et al. (2019a). Mir-449a-5p Mediates Mitochondrial Dysfunction and Phenotypic Transition by Targeting Myc in Pulmonary Arterial Smooth Muscle Cells. J. Mol. Med. 97, 409–422. 10.1007/s00109-019-01751-7 [DOI] [PubMed] [Google Scholar]
- Zhang C., Wang P., Mohammed A., Zhou Z., Zhang S., Ni S., et al. (2019b). Function of Adipose-Derived Mesenchymal Stem Cells in Monocrotaline-Induced Pulmonary Arterial Hypertension through Mir-191 via Regulation of Bmpr2. Biomed. Res. Int. 2019, 1–12. 10.1155/2019/2858750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu Y., Yan L., Wang S., Zhang M., Ma C., et al. (2019). Long Noncoding Rna Hoxaas3 Contributes to Hypoxia-Induced Pulmonary Artery Smooth Muscle Cell Proliferation. Cardiovasc. Res. 115, 647–657. 10.1093/cvr/cvy250 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang D., Li M., Plecitá-Hlavatá L., D’Alessandro A., Tauber J., et al. (2017). Metabolic and Proliferative State of Vascular Adventitial Fibroblasts in Pulmonary Hypertension Is Regulated through a Microrna-124/ptbp1 (Polypyrimidine Tract Binding Protein 1)/pyruvate Kinase Muscle axis. Circulation 136, 2468–2485. 10.1161/circulationaha.117.028069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Su H., Ma X., Xu X., Liang L., Ma G., et al. (2019). Mirna Let-7b Promotes the Development of Hypoxic Pulmonary Hypertension by Targeting Ace2. Am. J. Physiology-Lung Cell Mol. Physiol. 316, L547–L557. 10.1152/ajplung.00387.2018 [DOI] [PubMed] [Google Scholar]
- Zhang W.-F., Xiong Y.-W., Zhu T.-T., Xiong A.-Z., Bao H.-h., Cheng X.-S. (2017). MicroRNA Let-7g Inhibited Hypoxia-Induced Proliferation of PASMCs via G0/G1 Cell Cycle Arrest by Targeting C-Myc. Life Sci. 170, 9–15. 10.1016/j.lfs.2016.11.020 [DOI] [PubMed] [Google Scholar]
- Zhang W., Li Y., Xi X., Zhu G., Wang S., Liu Y., et al. (2020). MicroRNA-15a-5p I-nduces P-ulmonary A-rtery S-mooth M-uscle C-ell A-poptosis in a P-ulmonary A-rterial H-ypertension M-odel via the VEGF/p38/MMP-2 S-ignaling P-athway. Int. J. Mol. Med. 45, 461–474. 10.3892/ijmm.2019.4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shao R., Gao W., Sun G., Liu Y., Fa X. e. (2018). Inhibition of Mir-361-5p Suppressed Pulmonary Artery Smooth Muscle Cell Survival and Migration by Targeting Abca1 and Inhibiting the Jak2/stat3 Pathway. Exp. Cel Res. 363, 255–261. 10.1016/j.yexcr.2018.01.015 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen Y., Yao H., Lie Z., Chen G., Tan H., et al. (2019). Elevated Serum Circ_0068481 Levels as a Potential Diagnostic and Prognostic Indicator in Idiopathic Pulmonary Arterial Hypertension. Pulm. Circ. 9, 204589401988841. 10.1177/2045894019888416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Peng B., Han Y. (2018). Mir-23a Regulates the Proliferation and Migration of Human Pulmonary Artery Smooth Muscle Cells (Hpasmcs) through Targeting Bmpr2/smad1 Signaling. Biomed. Pharmacother. 103, 1279–1286. 10.1016/j.biopha.2018.04.172 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xu J. (2016). Mir-140-5p Regulates Hypoxia-Mediated Human Pulmonary Artery Smooth Muscle Cell Proliferation, Apoptosis and Differentiation by Targeting Dnmt1 and Promoting Sod2 Expression. Biochem. Biophysical Res. Commun. 473, 342–348. 10.1016/j.bbrc.2016.03.116 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Li Z., Wang Y., Wei L., Chen H. (2019). Overexpressed Long Noncoding RNA CPS1‐IT Alleviates Pulmonary Arterial Hypertension in Obstructive Sleep Apnea by Reducing Interleukin‐1β Expression via HIF1 Transcriptional Activity. J. Cel Physiol 234, 19715–19727. 10.1002/jcp.28571 [DOI] [PubMed] [Google Scholar]
- Zhao M., Chen N., Li X., Lin L., Chen X. (2019). Mir-19a Modulates Hypoxia-Mediated Cell Proliferation and Migration via Repressing Pten in Human Pulmonary Arterial Smooth Muscle. Life Sci. 239, 116928. 10.1016/j.lfs.2019.116928 [DOI] [PubMed] [Google Scholar]
- Zhou S., Jiang H., Li M., Wu P., Sun L., Liu Y., et al. (2019). Circular Rna Hsa_circ_0016070 Is Associated with Pulmonary Arterial Hypertension by Promoting Pasmc Proliferation. Mol. Ther. - Nucleic Acids 18, 275–284. 10.1016/j.omtn.2019.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Sun L., Cao C., Wu P., Li M., Sun G., et al. (2018). Hypoxia‐induced microRNA‐26b Inhibition Contributes to Hypoxic Pulmonary Hypertension via CTGF. J. Cel. Biochem. 119, 1942–1952. 10.1002/jcb.26355 [DOI] [PubMed] [Google Scholar]
- Zhou S., Zhang S., Wang Y., Yi S., Zhao L., Tang X., et al. (2015). Mir-21 and Mir-222 Inhibit Apoptosis of Adult Dorsal Root Ganglion Neurons by Repressing Timp3 Following Sciatic Nerve Injury. Neurosci. Lett. 586, 43–49. 10.1016/j.neulet.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Zhu T.-T., Sun R.-L., Yin Y.-L., Quan J.-P., Song P., Xu J., et al. (2019a). Long Noncoding Rna uca1 Promotes the Proliferation of Hypoxic Human Pulmonary Artery Smooth Muscle Cells. Pflugers Arch. - Eur. J. Physiol. 471, 347–355. 10.1007/s00424-018-2219-8 [DOI] [PubMed] [Google Scholar]
- Zhu T. T., Zhang W. F., Yin Y. L., Liu Y. H., Song P., Xu J., et al. (2019b). MicroRNA‐140‐5p Targeting Tumor Necrosis Factor‐α Prevents Pulmonary Arterial Hypertension. J. Cel Physiol 234, 9535–9550. 10.1002/jcp.27642 [DOI] [PubMed] [Google Scholar]