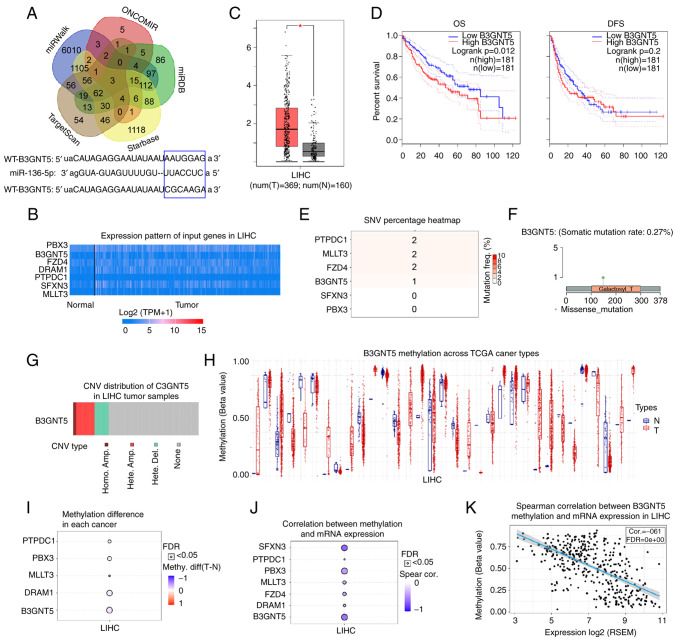
Figure 4.
Prediction and analysis of potential target genes of miR-136-5p. (A) Target genes of miR-136-5p were predicted using bioinformatics tools (starBase, miRWalk, TargetScan, ONCOMIR and miRDB). (B) Expression profile of seven target genes of miR-136-5p was analyzed using the UALCAN database. (C) Increased B3GNT5 expression was detected in LIHC in the GEPIA database. (D) OS and DFS of patients with liver cancer with different expression profiles of B3GNT5. GSCA database was used to study the SNV, copy number alteration and methylation levels of target genes. (E) SNV percentage of target genes in liver cancer. (F) Missense mutation of B3GNT5 in liver cancer. (G) CNV types of B3GNT5 in liver cancer. (H) B3GNT5 methylation levels across TCGA cancer types. (I) Methylation difference of target genes in liver cancer. (J) Correlation between methylation and mRNA expression levels of target genes of miR-136-5p. (K) Negative correlation between B3GNT5 methylation level and mRNA expression. *P<0.05. miR, microRNA; GEPIA, Gene Expression Profiling Interactive Analysis; OS, overall survival; DFS, disease-free survival; SNV, single nucleotide variation; CNV, copy number variation; TCGA, The Cancer Genome Atlas; WT, wild-type; MUT, mutant; FDR, false discovery rate; LIHC, liver hepatocellular carcinoma.